Choice of samples in extracellular microRNA research: Which fraction is better- exosomal or nonexosomal?
Abstract
MicroRNAs (miRNAs) are a species of noncoding RNA that post-transcriptionally regulates the expression of target mRNAs via degradation or translational repression. Recently, miRNAs in biofluids got extraordinary research priority in the field of noninvasive biomarker development because of their association with pathophysiological events, noninvasive way to obtain, and are markable stability in the extracellular harsh environment. Although several hypotheses have been proposed, the precise mechanisms of cellular release of extracellular miRNAs are not properly understood. In addition, it is often confusing to the researchers, which fraction of biofluid (based on release mechanisms) is suitable for biomarker development research. This review will briefly introduce extracellular miRNAs (EC miRNAs) and their release mechanisms. Furthermore, based on recent evidence, it will also summarize and suggest the appropriate fraction of biofluid to work with for biomarker development.
microRNAs AND EXTRACELLULAR microRNAs
microRNAs (miRNAs) are endogenously initiated noncoding RNA species of 18-24 nucleotide that post-transcriptionally regulate the expression of target mRNAs either via mRNA destabilization or transcriptional repression [1]. It is estimated that miRNAs cover 1-5% of the animal genome and bioinformatic studies predicted that almost 60% mammalian genes can be targeted by one or more miRNAs [2], thus they play a central role in cellular proliferation, migration, differentiation, survival, and cell fate. Since their discovery in the early 1990s [3], miRNAs have been broadly studied in different organism ranging from single- to multi-cellular organisms and found to be involved in important cellular pathways. According to the miRbase 21, more than 28000 miRNAs have been reported in all organisms that could potentially influence the expression of genes and associated with pathophysiological conditions [4].
While the majority of the miRNAs are detected intracellularly, a handful of miRNAs are found in various body fluids and commonly known as circulating miRNA or circulatory miRNA or extracellular miRNAs (ECmiRNAs). The first report of ECmiRNAs in biofluid was made by Chim and his colleagues [5] where they observed that placental miRNAs are readily detectable in maternal blood plasma. At the same time, another report claimed the existence of tumor-associated ECmiRNAs in the serum sample of the patients with diffused B-cell lymphoma [6]. The discovery of ECmiRNAs in blood serum/plasma opened a new era of miRNA research and a series of studies have consistently reported that the presence of ECmiRNAs in other biofluids including plasma [7], follicular fluid [8,9], amniotic fluid, and almost every other biofluids [10]. However, the total miRNA concentration, number of detected miRNAs, and level of detection vary significantly among different fluid types [10]. In addition, the expression profile of ECmiRNAs in certain biofluids in relation to different pathophysiological conditions indicates that ECmiRNAs could be selectively released from healthy cells [11–13] and has a great potential to serve as biomarkers to assess the pathological conditions.
CHARACTERISTICS OF ECmiRNAs
ECmiRNAs have distinct characteristics which are completely absent in cellular miRNAs. For instance, the introduction of cellular or synthetic miRNAs in plasma or any other fluid results in rapid degradation (within few minutes) and the addition of denaturing solution protect exogenous miRNAs from degradation by inactivating RNase activity in plasma [14]. In contrast, ECmiRNAs can stay for a long time in circulation or other biofluids without degradation and exhibit a higher level of resistance against the activity of high endogenous RNase indicating that ECmiRNAs avoid RNase activity by wrapping themselves with some protective manner [15]. While cellular miRNAs are susceptible to the unusual conditions like low or high pH, boiling, and prolonged storage time, ECmiRNAs showed remarkable stability under such conditions. In addition, ECmiRNAs are detectable in qRT-PCR after multiple freeze-thaw cycles (even up to 10 cycles) whereas cellular miRNAs start their signals after few freeze-thaw cycles [14,16]. In addition, Mitchell and his colleagues reported that a 24 h incubation of ECmiRNAs derived from serum at room temperature could not affect the specific expression patterns in qPCR [17]. In addition, Chen and colleagues reported that ECmiRNAs isolated from serum showed resistance against the treatment of RNase A, compared to other endogenous RNAs [15]. Majority of the serum derived ECmiRNAs are considerably expressed after being subjected to 3 h or overnight RNase treatment, on the other hand, large RNA species including 18s/28s rRNA, β-actin, GAPDH, and U6 degraded and showed no expression [15].
Although the release mechanism and remarkable stability of ECmiRNAs in bio-fluids are poorly understood, several hypotheses have been proposed to explain the mechanism. Nucleoprotein bound existence of RNAs is one of the earliest theory where the DNA fraction of the complex binds with RNA to form a DNA-RNA heterodimer [18]. Unexpectedly, a couple of years later, it has been reported that the protection and existence of RNA molecules in plasma or serum probably due to the binding with lipoprotein complex or lipid molecules, not with the DNA fraction of nucleoprotein [19].
MODE OF RELEASE OF ECmiRNAs
The mechanism of remarkable stability of ECmiRNAs in biofluids provoked us to speculate that these miRNAs are encapsulated by some manners to avoid degradation. Interestingly, exosome-mediated release, existence, and transport of miRNAs and mRNAs in cultured cells have reported shortly before the discovery of ECmiRNAs [20] which strongly suggests the hypothesis of the release of ECmiRNAs through extracellular vesicles. The membranes of extracellular vesicles are impermeable to RNase and this can keep the encapsulated miRNAs safe from degradation in the presence of high RNase activity in serum/plasma. Therefore, vesicles encapsulated release and existence of miRNAs in extracellular environment explained elegantly the remarkable stability of ECmiRNAs [21]. Later, Hunter and his colleagues reported that purified human peripheral blood microvesicles contain miRNAs [22] which further reinforce the theory of microvesicles encapsulated miRNAs in the extracellular environment.
Extracellular vesicles are a combination of a heterogeneous population of exosomes and microparticles (shedding vesicles). Exosomes are homologous small vesicles (50-90 nm) [23], whereas microparticles/microvesicles (MVs) are lipid vesicles that are <1 μm in diameter and are secreted into extracellular environment by different types of cells and platelets [24]. While exosomes are generated via endocytic recycling pathway (inward budding), MVs are produced from plasma membrane via outward budding and fission. Thus, the membrane composition of MVs is much similar to the plasma membrane of their parent cells. Hence the membrane composition of MVs is predominantly depended on the cell of origin [25]. In the following years, several studies reported the existence of miRNAs in various biofluid in association with either exosomes [12,23] or microvesicles [26,27].
In 2011, the vesicle-enwrapped release and existence of miRNAs were hugely challenged when independent two groups claimed that a large fraction of ECmiRNAs exists in the cell culture media [14] and in human plasma [28] that are outside of shedding vesicles and exosomes. In addition, authors claimed that only 10% of ECmiRNAs are released in plasma through vesicles and potentially 90% of them are associated with protein complexes [28]. Furthermore, when size-exclusion chromatography was used to complete separation of vesicles from protein complex, it showed that most of the ECmiRNAs are ribonucleoprotein complex associated and only a few miRNAs are predominantly associated with microvesicles [28]. It is important to note that all mature miRNAs become associated with the AGO protein families during their intracellular synthesis process [2]. Therefore, it is logical to think that ECmiRNAs could exist with AGO protein, particularly AGO2 protein, in the extracellular environment. In addition, it has been known for a long time that AGO2 proteins are remarkably stable in nuclease- and protein-rich environment which also explained the stability of protein complex associated ECmiRNAs [14]. Later, Gallo and colleagues reported that several miRNAs including miR-16 and miR-92a were detected in the pellet of ultracentrifuges serum samples while they are barely detected in the supernatant [29]. It was unexpected because Arroyo and colleagues reported that miR-16 and miR-92a were predominantly associated with nucleoprotein [28]. The difference in the results of two groups may be due to the differences in the sample processing, isolation, and data normalization method.
In the following years of ECmiRNA discovery, accumulated studies have reported that ECmiRNAs are packaged into apoptotic bodies[30], high-density lipoprotein (HDL) particles [31], microvesicles [22], exosomes [23], and AGO2 proteins [28]. A pictorial representation of release and existence of extracellular miRNAs in biofluids are presented in Figure 1. The inclusion of apoptotic body associated miRNAs in ECmiRNA category is questionable due to its size (comparable with the size of cellular debris and platelets). In addition, in the commonly used protocol for ECmiRNA isolation, during the sample processing step, apoptotic bodies are removed with cell debris in the initial ultracentrifugation step. Thus, this fraction of ECmiRNAs is overlooked and understudied.
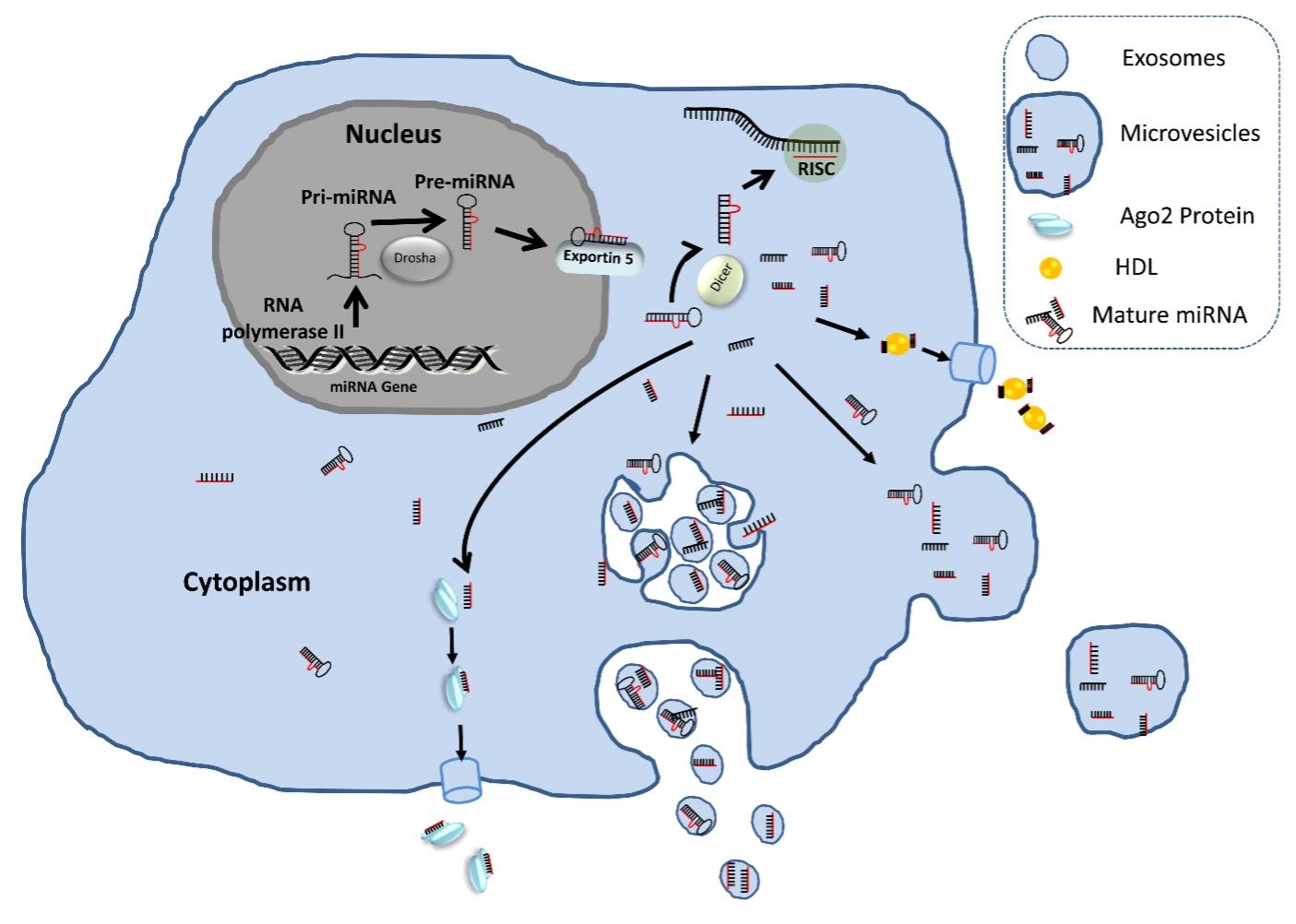
CHOICE OF SAMPLES FOR ECmiRNA RESEARCH
Soon after the discovery of ECmiRNAs, they received an utmost research priority due to their potential as a noninvasive biomarker and to understand the disease pathogenesis. Therefore, much of the efforts in profiling ECmiRNAs in various biofluid is involved in investigating the expression patterns of ECmiRNAs and correlates them with different physiological status and disease progression. Despite the widespread interests in developing biomarkers based on ECmiRNAs, heterogeneity among the results in different studies have been observed and hence the use of ECmiRNAs as reliable and reproducible biomarker remain in infancy [4]. The contributing factors in the variation in results can be both preanalytical and analytical. Preanalytical factors are starting material, methods of collection, processing, RNA extraction, and expression analysis. The source and composition of these biofluids are quite different; therefore, the processing and isolation methods need to be changed/adopted according to the origin of biofluids. Thus, to avoid the variation in the results of ECmiRNA research, a standardized protocol should be followed from sample collection to data analysis [4].
One of the major points to be considered in ECmiRNA research is the choice of sample or starting materials. For instance, when ECmiRNAs are quantified from circulation, a critical decision should be made that is which fraction will be the starting material- whole blood or sera or plasma? Essentially, we must discard whole blood from the list as it contains cellular fractions (white and red blood cells) [32]. The basic difference between serum and plasma is the absence or presence of clotting factors, respectively. Both plasma and serum are used for ECmiRNA research. If plasma is the starting choice, ethylenediaminetetraacetic acid (EDTA) is the better choice over heparin as it is known as PCR inhibitor. Although many studies investigated plasma and serum side-by-side and found no or little difference in ECmiRNA quantification [15], consistently higher concentrations were found in serum samples [33]. Similarly, cellular contribution in other biofluids should be discarded before subjected to ECmiRNA expression analysis. This could be achieved by using a centrifugation step at 25,000 × g for 30 min [12]. In addition to cellular fraction, the centrifugation step removes the cellular debris, apoptotic bubbles, and fractions of large macroparticles.
Now, in the cell-free centrifuged biofluid, ECmiRNAs can be existed by two means such as exosomal and nonexosomal (includes microvesicles, AGO2 protein, and HDL).When AGO2 protein-coupled ECmiRNAs are kept for several weeks in cell lysates without protease inhibitors, it showed no degradation. This clearly suggests that the protein complex associated ECmiRNAs may simply derive from the passive release of cytosolic components during cell death [14]. HDL and LDL can carry a pure population of ECmiRNA in biofluids [31] and their functional relevance has been demonstrated in themodel organism [34] and human [35]. However, it is not entirely clear that HDL coupled ECmiRNAs are selectively and actively released from the donor cells and spontaneously taken up by the recipient cells. Therefore, it is clear that nonexosomal ECmiRNAs are the result of nonspecific and passive release from the cells and may not be the better choice for the biomarker study.
Exosome formation and release is a tightly coordinated and complex process which is orchestrated by enzymatic activation and energy in the form of adenosine triphosphate (ATP) [36], and the expression patterns of exosomal ECmiRNAs(Exo-miRs)obtained from culture media and various bio-fluids derived from bio-fluids are different from their cell of origin suggesting there might be an active selection mechanism for exosome release and their contents. In contrast to nonexosomal ECmiRNAs, exosomal ECmiRNAs (Exo-miRs) are released from viable cells and can be taken up by the recipient cells and modulate the expression of genes [12] and trigger functional effects [20,37], however, the sorting and secretion of Exo-miRs are not fully understood. Pegtel and colleagues demonstrated the potential of Exo-miRs to facilitate viral infection [37] while Sohel and colleagues showed the gene expression modulation capacity of follicular fluid derived Exo-miRs in the recipient cells [12]. Many other studies have further indicated that miRNAs can be transported to the recipient cells via exosomes and could facilitate the cell-to-cell communication in vitro [20,38] and in vivo [39]. Exo-miRs are particularly important in cancer biomarker development. The main reason behind this is- cancer cells, in a given microenvironment, constitutively secrete exosomes to mediate cell-cell communication with same neighboring cells (autocrine manner), with different neighboring cells (paracrine manner), and with distant cells (endocrine manner) [40]. Therefore, much of the efforts were made in developing cancer biomarkers using Exo-miRs whereas nonexosomal ECmiRNAs were overlooked.
ECmiRNAs are the new celebrity of miRNA research where Exo-miRs dominating the field. The specific expression pattern of Exo-miRs in various biofluids is distinctly associated with various pathophysiological conditions. Thus, Exo-miRs have the great potential in biomarker development and perhaps, in future, Exo-miRs will be routinely used for therapeutic purpose. Indeed, among all ECmiRNAs, Exo-miRs are released from viable cells and have the ability to modulate the biological function in recipient cells, thus, the exosomal fraction of biofluid is always beena better choice over nonexosomal fraction for biomarker development and/or therapeutic study.
CONFLICT OF INTEREST
The author declares that no conflict of interest exists.
FUNDING
This research does not receive any fund from public or privet organization.
References
- [1]Tesfaye D, Salilew-Wondim D, Gebremedhn S, Sohel MMH, Pandey HO, Hoelker M et al. Potential role of microRNAs in mammalian female fertility. Reprod Fertil Dev. 2017; 29: 8–23.
- [2]Hossain MM, Sohel MMH, Schellander K, Tesfaye D. Characterization and importance of microRNAs in mammalian gonadal functions. Cell Tissue Res. 2012; 349: 679–90.
- [3]Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–54.
- [4]Sohel MMH. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev Life Sci 2016; 10: 175–186.
- [5]Chim SSC, Shing TKF, Hung ECW, Leung T-Y, Lau T-K, Chiu RWK et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 2008; 54: 482–90.
- [6]Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008; 141: 672–675.
- [7]Noferesti SS, Hoelker M, Sohel MMH, Salilew-Wondim D, Rings F, Schellander K et al. Effect of ovarian hyperstimulation on circulatory microRNA profile in bovine follicular fluid and blood plasma. Reprod Fertil Dev 2012; 25: 251.
- [8]Sohel MMH, Salilew-Wondim D, Hölker M, Rings F, Schellander K, Tesfaye D. Circulatory microRNA signatures in follicular fluid in relation to the growth status of bovine oocytes. Reprod Fertil Dev 2012; 25: 251.
- [9]Sohel MMH, Noferesti SS, Salilew-Wondim D, Hoelker M, Rings F, Tholen E et al. Relative abundance of extra-cellular miRNAs in bovine follicular fluid: Implication for cell–cell communication during oocyte growth. Anim Reprod Sci 2014; 149: 98.
- [10]Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010; 56: 1733–41.
- [11]Pigati L, Yaddanapudi SCS, Iyengar R, Kim D-J, Hearn SA, Danforth D et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One 2010; 5: e13515.
- [12]Sohel MMH, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft C et al. Exosomal and non-exosomal transport of extra-cellular microRNAs in follicular fluid: Implications for bovine oocyte developmental competence. PLoS One 2013; 8. doi:10.1371/journal.pone.0078505.
- [13]Noferesti SS, Sohel MMH, Hoelker M, Salilew-Wondim D, Tholen E, Looft C et al. Controlled ovarian hyperstimulation induced changes in the expression of circulatory miRNA in bovine follicular fluid and blood plasma. J Ovarian Res 2015; 8: 81.
- [14]Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011; 39: 7223–33.
- [15]Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006.
- [16]Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008; 110: 13–21.
- [17]Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105: 10513–8.
- [18]Sisco KL. Is RNA in serum bound to nucleoprotein complexes? Clin Chem 2001; 47: 1744–5.
- [19]El-Hefnawy T, Raja S, Kelly L, Bigbee WL, Kirkwood JM, Luketich JD et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem 2004; 50: 564–73.
- [20]Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654–9.
- [21]Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci 2012; 37: 460–5.
- [22]Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 2008; 3: e3694.
- [23]Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int 2010; 78: 838–48.
- [24]Aharon A, Katzenell S, Tamari T, Brenner B. Microparticles bearing tissue factor and tissue factor pathway inhibitor in gestational vascular complications. J Thromb Haemost 2009; 7: 1047–50.
- [25]Lee Y, El Andaloussi S, Wood MJA. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet 2012; 21: R125-34.
- [26]Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One 2009; 4: e4722.
- [27]Castellana D, Kunzelmann C, Freyssinet J-M. Pathophysiologic significance of procoagulant microvesicles in cancer disease and progression. Hämostaseologie 2009; 29: 51–7.
- [28]Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011; 108: 5003–8.
- [29]Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012; 7: e30679.
- [30]Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal 2009; 2: ra81.
- [31]Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011; 13: 423–433.
- [32]Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012; 5: 492–7.
- [33]Wang K, Yuan Y, Cho J-H, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One 2012; 7: e41561.
- [34]Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun 2014; 5: 3292.
- [35]Niculescu LS, Simionescu N, Sanda GM, Carnuta MG, Stancu CS, Popescu AC et al. MiR-486 and miR-92a Identified in Circulating HDL Discriminate between Stable and Vulnerable Coronary Artery Disease Patients. PLoS One 2015; 10: e0140958.
- [36]Yáñez-Mó M, Siljander PR-M, Andreu Z, Zavec AB, Borràs FE, Buzas EI et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell vesicles 2015; 4: 27066.
- [37]Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MAJ, Hopmans ES, Lindenberg JL et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 2010; 107: 6328–33.
- [38]Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan MLG, Karlsson JM et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012; 119: 756–66.
- [39]da Silveira JC, Veeramachaneni DNR, Winger QA, Carnevale EM, Bouma GJ. Cell-Secreted Vesicles in Equine Ovarian Follicular Fluid Contain miRNAs and Proteins: A Possible New Form of Cell Communication Within the Ovarian Follicle. Biol Reprod 2012; 86: 71–71.
- [40]Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp Mol Med 2017; 49: e285–e285.