Biochemical and molecular identification of antibiotic-producing bacteria from waste dumpsite soil
Abstract
Antibiotics are the secondary metabolites produced by bacteria and fungi to defend themselves from other pathogens. These secondary metabolites are being produced and used as a drug to cure different diseases. However, antibiotic resistance is a common problem that demands an urgent need to discover new antibiotics routinely. Several approaches have been performed to develop novel and potent antibiotics from natural sources against pathogenic bacteria. Among the different sources soil has been considered as the potent natural source of obtaining bacteria with the ability to produce novel antibiotics. The present work has been focused on the isolation of antibiotic producing bacteria from the soil samples collected from waste dumpsite. Among the 5 microbial isolates, 2 were shown to have inhibitory activities against Escherichia coli and Salmonella paratyphi. Morphological and biochemical tests revealed that both strains were Bacillus species with some differences in cultural characteristics. Molecular identification was performed by sequencing of the amplified 16S rRNA PCR products. The result showed that the two microbes were Bacillus subtilis and Bacillus cereus with 98% and 97% similarity score, respectively. This study suggests that Bacillus species have the potential to produce antibiotics against a broad spectrum of microbial growth and will be helpful in improving these strains for better production.
INTRODUCTION
Microorganisms produce different secondary metabolites, known as antibiotics to maintain their territory from other microorganisms. Antibiotics are used for the cure of diseases of humans [1]. After the discovery of antibiotics it has revolutionized the field of medicine [2]. However, the misemployment or immense use of it is responsible for drug resistant property of the relevant pathogens [3, 4]. The ratio of resistant bacteria is increasing and it is threatening treatment methods of modern medicine [5]. So, there is enormous importance of searching new antibiotics that can kill or destroy the resistant bacteria.
There are several key reasons of becoming a drug resistant as available source of antibiotics, their immense usage and random disposal. Therapeutic uses of the antibacterial products in human, animal and agriculture also favour the survival and migration of resistant bacteria [6]. The improper uses of animal wastes as fertilizers are also responsible for spreading of resistance bacteria in soil. Consumption of animal products is the way of transmission of drug resistant bacteria to human [7]. It is a situation demand to develop new, effective and safe antibiotics to combat the possible danger of drug-resistant pathogens.
The microbial secondary metabolites are considered as the most promising resources for the novel antibiotics [8, 9]. Antibiotic producing bacteria are abundant in soil where they produce antimicrobial metabolites with specific activities against co-existing microorganisms [10]. The antibiotic diversities in the soil depend on the availability of nutrient, soil moisture, temperature and pH [4]. It was reported earlier that among the all antibiotic producing bacteria, Bacillus species are the most abundant in soil. They can adapt very easily in any habitat by their ability to form spores to survive a range of adverse environments [11]. To identify the diversified physiology of Bacillus species, elaborate biochemical tests are required. Molecular identification techniques such as 16S rRNA and gyrase B sequence analysis are considered as promising methods for phylogenetic analysis of bacteria [11]. In the present study, we aimed to isolate and characterized antibiotic producing bacteria from waste dumpsite soil in Khulna city, Bangladesh.
MATERIALS AND METHODS
Collection of samples
Soil samples were collected from waste dumpsites in Khulna city, Bangladesh. After removing all surface debris the site was dug into 4-5 cm and approximately 10-15 g of the soil was collected in a sterile plastic bag using a sterile spatula. Five different sites of the dumping area were used to collect soil samples. The bag contained the soil samples were labelled specifically, transported into laboratory and stored at 4 0C till further analysis.
Isolation and maintenance of soil bacteria
Soil bacteria were isolated by the well-known ‘Crowded plate’ technique following standard serial dilution. One gram of each soil sample was weighed and soaked in 10 ml of sterile distilled water to get 1:10 dilution. After vigorous shaking it was allowed to settle for sediment. The supernatant was collected and then serially diluted. An amount of 100 µl from each dilution was aseptically spread on labelled nutrient agar plates using a glass rod. After few seconds the agar plates were incubated at 37 oC for 24 to 48 hrs. The distinguishable colonies were selected for streaking on agar plate separately to gain pure colonies. Pure culture was stored at 4 °C for subsequent studies.
Physiological characterization of the isolated bacteria
The morphology of each colony of the bacterial isolates on agar plates was observed under microscope. After an incubation of 24 hrs at 37 ºC, individual colonies were characterized based on their color, shape, appearance, colony diameter and transparency. Gram staining (Thermo Fisher Scientific, Massachusetts, USA) method was used to distinguish between gram +ve and gram –ve bacteria.
Secondary screening of antibiotic producing bacteria
The pure colonies from different bacterial isolates were tested for antibacterial activity against two gram negative bacteria, Escherichia coli and Salmonella paratyphi. The rationale of choosing gram negative bacteria is that they possess endotoxin and have a history of serious infections in humans. The agar diffusion method was used for secondary screening of antibiotic producing bacteria. An amount of 100 µl of the pathogenic culture was inoculated in each of the plates by spread plate technique. The overnight culture of the colonies isolated was centrifuged and the supernatant (250µl) was absorbed on the discs for final use. These discs were placed on the agar plates and then kept in the incubator overnight. Blank disc were used as a negative control. The diameter of each inhibited zone was measured and evaluated the intensity of antagonism against test pathogens.
Biochemical screening of the isolated bacteria
The bacterial isolates were characterized biochemically to evaluate their chemical nature. We conducted oxidase test, catalase test, Voges-Proskauer test, methyl red test, indole production test, starch hydrolysis test, citrate utilization test, carbohydrate fermentation tests and growth on MacConkey agar according to the standard protocols.
Extraction of genomic DNA from selected bacteria
The bacterial broth was centrifuged to discard supernatant for cell pellet. The pellet was washed with 0.9% saline and suspended in the digestion buffer. The genomic DNA extraction process was performed using automated DNA extractor (Invent Technologies Ltd., Dhaka, Bangladesh) according to the manufacturer instructions. The concentration of isolated DNA was measured using spectrophotometer at 260 and 280 nm of wavelength. The purity of genomic DNA was checked before further use.
Molecular identification of bacteria by 16S rRNA amplification and sequencing
The 16S rRNA gene fragments were amplified using GoTaq® Green Master Mix (Promega Corporation, Wisconsin, USA) according to the manufacturer instruction. The universal primers were used as forward (5’-AGA GTT TGA TCM TGG CTC AG-3’) and reverse (5’- CGG TTA CCT TGT TAC GAC TT-3’) primer. In brief, the PCR started with an initial denaturation for 3 min at 95 °C. The optimum denaturation was performed 32 cycles at 95 °C for 30 s for each cycle. The annealing temperature was 55 °C for 30 s followed by an extension at 72 °C for 1 min. The PCR was ended with the final step of extension for 5 min at 72 °C. The amplified PCR products were run on 1% agarose gel in the electrophoresis tank. The gel was stained in ethidium bromide and scanned using a trans-illuminator AlphaImager (Alpha Innotech, California, USA).
The amplified gene fragments were purified and sequenced from Invent Technologies Ltd., Dhaka, Bangladesh. 16S rRNA gene sequences were aligned using a sequence alignment editor, BioEdit 7.2 and exported into BLAST to identify matches with existing reference sequences.
RESULTS
Crowded plate technique screened out distinct isolates
Colonies with distinguished morphology were found when soil samples cultured on media following crowded plate technique. Plates with crowd but well demarcated colonies were selected for screening. Five different colonies at dilution 104 showed distinguishable properties. Hence, these 5 antagonistic bacterial colonies (BI-1, BI-2, BI-3, BI-4 and BI-5) were isolated and their pure cultures were stored at 4 oC for further use.
Secondary screening of different isolates
The 5 bacterial isolates were tested for their antibiotic efficiency by disc diffusion assay against the pathogenic test strains Escherichia coli and Salmonella paratyphi. The result showed that except BI-5 all other isolates have antibacterial activity. BI-2, BI-3 and BI-4 showed prominent zone of inhibition against E. coli whereas, BI-1, BI-2, BI-3 and BI-4 showed zone of inhibition against S. paratyphi (Table 1). The best two isolates (BI-2 and BI-3) were selected for further analysis.
Table 1. Antagonistic activity of bacterial isolates against test pathogens.
Morphological characterization of BI-2 and BI-3
Analysis of the colony morphology on agar plate is the conventional method used to characterize bacterial growth. Microscopic examination of BI-2 and BI-3 revealed circular shaped colonies with smooth surfaces. The detailed characteristics of their colony morphology are presented in Table 2. Both of the isolates were grown at the surface of the broth, so the growth in broth was pellicle. This result indicated that they were aerobic. Furthermore, gram staining technique was applied to identify their cell wall properties. Result showed that both isolates retained purple color which denoted them as gram positive bacteria. The microscopic observation of BI-2 and BI-3 suggested that their shape were bacilli in nature (Figure 1). Several biochemical tests were conducted to confirm the identity of isolated strains which are listed in Table 3. The above results suggested that BI-2 and BI-3 belong to the gram positive Bacillus species.
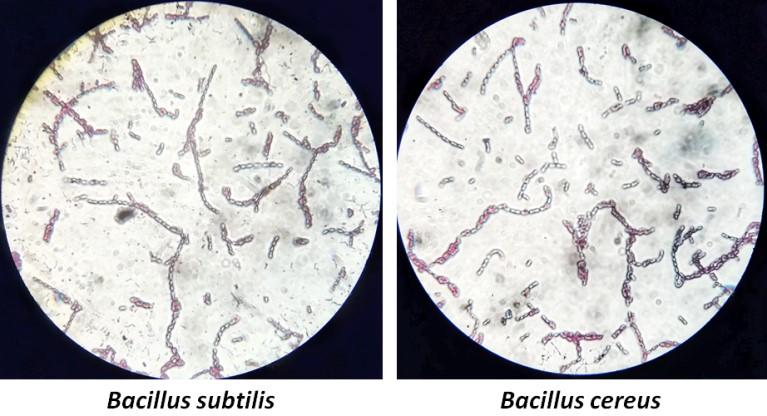
staining. The rod shaped bacilli are clearly visible with endospores.
Table 2. Colony morphology and gram staining of bacterial isolates.
Table 3. Biochemical test results for the identification of bacterial isolates.
Molecular identification of BI-2 and BI-3
The PCR products of bacterial 16S rRNAs were purified and sequenced. The sequence of bacterial isolates BI-2 and BI-3 were subjected to NCBI BLAST. The bacterial isolate BI-2 showed 98% of similarity with the available database sequence of Bacillus subtilis in BLAST. On the other hand, isolate BI-3 showed 97% similarity with Bacillus cereus. The sequences of BI-2 and BI-3 were deposited in GenBank and the accession numbers were MK073920.1 and MK074711.1, respectively. The detailed sequences of BI-2 and BI-3 were presented in Table 4. The phylogenetic positions of the isolates were also evaluated by constructing a phylogenetic tree using neighbor-joining methods of MEGA7 software [12] (Figure 2).
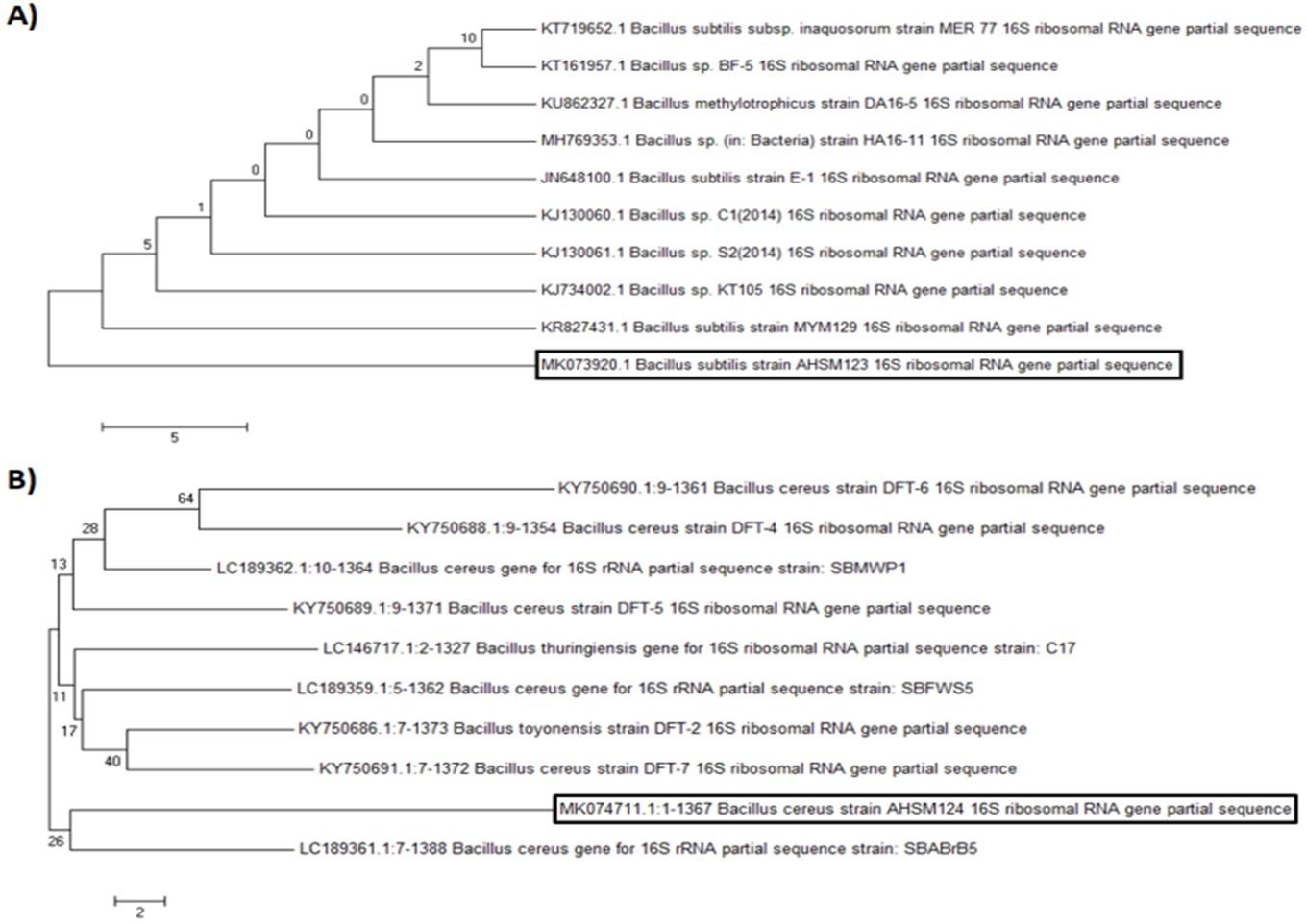
Table 4. 16S rRNA gene sequence of antagonistic bacterial isolates BI-2 and BI-3.
DISCUSSION
Soil is one of the best sources for isolating novel antibiotics as many scientists have chosen it for their research [13-16]. It was reported that the heterogeneity of soil environment results heterogeneous population of soil bacteria [17]. In this present study, isolation of the antibiotic producing microorganisms was performed from waste dumpsite soil. We have isolated two antibiotic producing bacteria and submitted their 16S rRNA sequences in the NCBI BLAST.
Morphological identification was done by observing the colony morphology and by gram staining. Gram staining method is widely used and unique conventional method of bacterial characterization [9, 18]. The staining result showed that both of the bacterial isolates BI-2 and BI-3 were gram positive in nature. It was reported that most of the soil isolates are gram positive in nature [14] which supports our result.
During secondary screening the bacterial culture filtrate was used to observe the antagonistic property of BI-2 and BI-3. The use of bacterial culture filtrate in agar diffusion method is a common practice among scientists [19, 20]. The secondary screening of the isolates BI-2 and BI-3 was done against the human pathogens, E. coli and S. paratyphi. The maximum zone of inhibition was shown by BI-2 against E. coli. E. coli is one of the common pathogenic bacteria in the soil. Recently the antibiotic resistant pathogenic E. coli were isolated from household soil [21]. Additionally, they found that 42.3%, 12.6% and 10% E. coli isolates were single antibiotic resistant, multidrug resistant and potentially pathogenic, respectively. Hence, our bacterial isolate BI-2 would be potential against pathogenic E. coli.
For molecular characterization of bacterial isolates 16S rRNA amplification and sequencing was performed. The 16S rRNA gene sequencing was considered as the improved method for identification of bacteria from different samples [22]. The sequences of bacterial isolates BI-2 and BI-3 were matched with Bacillus subtilis and Bacillus cereus, respectively. It was reported that Bacillus species were one of the dominant bacterial species in soil [23, 24]. Gram positive bacteria are more susceptible to antibiotics than gram negative bacteria for their composition of outer surface. We observed bigger inhibited zones against E. coli and S. paratyphi which indicated the nobility of our isolated strains.
One of the major reasons to be antibiotic-resistant is the excessive use of it as a drug. There is a consistent need for new antibiotics that would be effective against different pathogens. The natural products are the major sources of secondary metabolites in nature. Hence, we aimed to isolate and characterize soil bacteria with antibiosis activity. We have identified Bacillus subtilis and Bacillus cereus from waste dumpsite soil which has antibacterial activity against E. coli and S. paratyphi, respectively. We hope that our findings will help further the commercial production of new antibiotic drugs after proper down-stream processing.
ACKNOWLEDGEMENT
This research was partially funded by Khulna University Research Cell; grant number KURC-RGP-13/2018. The authors are grateful to Laxmon Chandra Roy for his invaluable technical assistance.
AUTHOR CONTRIBUTIONS
AH was involved in the conception and design of the experiments. TT and MJS contributed to perform the experiments and also analyzed data. CM contributed to drafting the article and contributed to revising it critically for important intellectual content. AH made the final approval of the version to be published.
CONFLICTS OF INTEREST
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- [1]Mowafy EI, Attia M, Shaaban MT, Turky AS, Awad NM. Ecological studies on microorganisms producing antimicrobial agents from different soil types. Res J Pharm Biol Chem Sci. 2015; 6(5): 1020-30.
- [2]Wohlleben W, Mast Y, Stegmann E, Ziemert N. Antibiotic drug discovery. Microb Biotechnol. 2016; 9(5): 541-8.
- [3]Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015; 40(4): 277-83.
- [4]Nasfi Z, Busch H, Kehraus S, Linares-Otoya L, Konig GM, Schaberle TF et al. Soil bacteria isolated from Tunisian arid areas show promising antimicrobial activities against gram-negatives. Front Microbiol. 2018; 9: 2742.
- [5]Frieri M, Kumar K, Boutin A. Antibiotic resistance. J Infect Public Health. 2017; 10(4): 369-378.
- [6]Gebreyohannes G, Moges F, Sahile S, Raja N. Isolation and characterization of potential antibiotic producing Actinomycetes from water and sediments of Lake Tana, Ethiopia. Asian Pac J Trop Biomed. 2013; 3(6): 426-35.
- [7]Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011; 24(4): 718-33.
- [8]Yunus FN, Khalid ZZ, Rashid F, Ashraf A, Iqbal MN, Hussain F. Isolation and Screening of Antibiotic producing Bacteria from soil in Lahore City. PSM Microbiology. 2016; 1(1): 01-04.
- [9]Kumar S, Chaurasia P, Kumar A. Isolation and characterization of microbial strains from textile industry effluents of Bhilwara, india: analysis with bioremediation. J Chem Pharm Res. 2016; 8(4): 143-150.
- [10]Cavalcanti, Maria AQO, Luciana GF, Maria JL, Débora M. Filamentous fungi isolated from soil in districts of the Xingó region, Brazil. Acta Botanica Brasilica. 2006; 20: 831-837.
- [11]Parvathi A, Krishna K, Jose J, Joseph N, Nair S. Biochemical and molecular characterization of Bacillus pumilus isolated from coastal environment in Cochin, India. Braz J Microbiol. 2009; 40(2): 269-75.
- [12]Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33(7): 1870-4.
- [13]Nike AR, Hassan SA, Ajijolakewu AFB. Soil screening for antibiotic – producing microorganisms. Adv Environ Biol. 2013; 7(1): 7-11.
- [14]Wadetwar RN, Patil AT. Isolation and characterization of bioactive Actinomycetes from soil in and around Nagpur. Int J Pharm Sci Res. 2013; 4: 1428-1433.
- [15]Kaur S, Kaur J, Pankaj PP. Isolation and characterization of antibiotic producing microorganisms from soil samples of certain area of Punjab region of India. Int J Pharm Clin Res. 2014; 6: 312-315.
- [16]Gislin D, Sudarsanam D, Antony Raj G, Baskar K. Antibacterial activity of soil bacteria isolated from Kochi, India and their molecular identification. J Genet Eng Biotechnol. 2018; 16(2): 287-294.
- [17]Constancias F, Terrat S, Saby NP, Horrigue W, Villerd J, Guillemin JP et al. Mapping and determinism of soil microbial community distribution across an agricultural landscape. Microbiologyopen. 2015; 4(3): 505-17.
- [18]Diddi K, Chaudhry R, Sharma N, Dhawan B. Strategy for identification & characterization of Bartonella henselae with conventional & molecular methods. Indian J Med Res. 2013; 137(2): 380-7.
- [19]Yoshida S, Hiradate S., Tsukamoto T, Hatakeda K, Shirata A. Antimicrobial activity of culture filtrate of Bacillus amyloliquefaciens RC-2 isolated from Mulberry leaves. Phytopathology. 2001; 91(2): 181-7.
- [20]Sheikh HMA. Antimicrobial activity of certain bacteria and fungi isolated from soil mixed with human saliva against pathogenic microbes causing dermatological diseases. Saudi J Biol Sci. 2010; 17(4): 181-7.
- [21]Montealegre MC, Roy S, Boni F, Hossain MI, Navab-Daneshmand T, Caduff L et al. Risk factors for detection, survival, and growth of antibiotic-resistant and pathogenic Escherichia coli in household soils in rural Bangladesh. Appl Environ Microbiol. 2018; 84(24): e01978-18.
- [22]Lau SKP, Teng JLL, Ho CC, Woo PCY. Gene amplification and sequencing for bacterial identification. Methods in Microbiology. 2015; 42: 433-464.
- [23]Singh AP, Singh RB, Mishra S. Studies on isolation and characterization of antibiotic producing microorganisms from industrial waste soil sample. Open Nutraceuticals J. 2012; 5(1): 169-173.
- [24]Abbas S, Senthilkumar R, Arjunan S. Isolation and molecular characterization of microorganisms producing novel antibiotics from soil sample. Eur J Experiment Biol. 2014; 4(5): 149-155.