Presumptive correlation between phenotypic, genotypic and symbiotic diversities with antibiotic susceptibility traits of rhizobial strains from plant legumes
Abstract
This investigation was carried out to study the extremely diversified characteristics of rhizobial strains isolated from locally produced legumes of different varieties. The rhizobial isolates designated as S 1, S 2, S 3, S 4, S 5 and S 6 were purified and subjected to various morphological, cultural, antibiogram and biochemical and plasmid profiling to unmask their diversified nature to some extent. The study revealed that the Sesbania strain (S 2) possessed extraordinary properties for most other strains. Though the strain is allocated in the slow-growing group, it showed an atypical fast growth rate, and also it produced a profuse amount of exopolysaccharide (EPS)/ lipopolysaccharide (LPS). The Arachis and Vigna strains also showed a faster growth rate than usual, whereas the rate of EPS/LPS production was profuse in the cases of Vigna and Lens strains. This rate was noted low in cases of Pisum and Arachis strains, while Phaseolus produced a moderate amount of EPS/LPS. Like Sesbania, the Pisum strain also absorbed Congo red dye strongly and including Sesbania, all the other strains ware catalase-negative, which was an unusual property of rhizobial strains. Antibiogram profile of the test strains was done to retain experimental strains after plant infection test, as overlapping host range is a common phenomenon in symbiosis. The antibiogram profiles showed that more or less all the strains were resistant to Ampicillin, Bacitracin, and Amoxicillin. Phaseolus strain was intermediate to Bacitracin, and Pisum strain was susceptible to Ampicillin, which can be related to loss of the plasmid. In plasmid profiling of the strains, several numbers of plasmid DNA band was observed under UV transilluminator after gel electrophoresis. In the present study, rhizobial species from different leguminous plants were detected, and plasmids were cured of the cell using acridine orange to determine their effect on Rhizobium spp. for antibiotic resistance. The findings of the research point out the diverse nature of rhizobial isolates with specific reference to the strains infective on Sesbania aculeate, which poses problems to characterize strains infective of different legumes.
INTRODUCTION
Protein and thus amino acids are the units of life. So, the amino acids are called the “building blocks of life” [1, 31]. Nitrogen (N) is a must component for all amino acids. Thus, all plants and animals must have nitrogen for growth. The air we breathe is 78% nitrogen gas and 21% oxygen. There is ~35,000 tons of free N above every acre of land, but this gaseous form is unavailable to plant or animal life [2, 23-25, 32]. Fortunately, nature has provided us with a simple and cheap method of obtaining some of this N from the atmosphere by growing legumes. Approximately 110 million tons of N is required for the world’s annual food production but only 7 million tons are supplied by the fertilizer industry; the rest come from legumes [25].
Legumes are plants, like peas, beans, soybean, alfalfa, clover, and aeschynomene, which have special bacteria in their rooting system and make use of gaseous nitrogen from the air [2, 3, 31]. This phenomenon is called biological nitrogen fixation (BNF). In 1888 Hellriegelg and Wilfrath provided a major scientific contribution by establishing a rational explanation of this phenomenon. This pioneering work laid the foundation for all subsequent studies in nitrogen fixation and the legume-bacteria symbiosis [13, 14, 23-25, 31]. The agricultural importance of nitrogen fixation is not only to provide ammonium to the crops, but the independence from using nitrogenous fertilizer also minimizes pollution of water tables, lakes, and rivers. The legume-rhizobium symbiosis and the corresponding physiological adaptations also provide a convenient model for studying aspects of plant-microbe interactions and evolution [3, 23-25, 31]. Many legumes are important nutritional crops for human and animal consumption. In Bangladesh, like most other third world developing countries, malnutrition is a fact that still is resisting healthy life and improvement of lifestyle. The main food crops in Bangladesh are the non-nodulated rice and wheat. The protein content of these crops is not sufficient to improve the situation of malnutrition in this country [9, 10, 23-25]. Hence the long-standing and ambitious goal of research in biological nitrogen fixation has been to extend the nitrogen-fixing symbiosis to non-nodulated cereal plants such as rice and wheat. In the developing countries, recent advantages in symbiotic Rhizobium-legume symbiosis at a molecular level and the discoveries of entophytic associations will help in tracking this tremendous task [15, 25, 31].
With the increase of intensive cultivation, our soils are becoming less fertile day by day, particularly with the depletion of nitrogen-the major limiting factor for growth. Legumes besides being protein sources, add nitrogen and other nutrients to the land [2]. Grain legumes provide valuable nutritious seeds, and when effectively nodulated, can supplement nitrogen where cereals and other non-leguminous crops are grown. Previous studies have shown that the Rhizobium genus members posed an amazing diversity of nature [16, 23]. The diversity of their ability to inhabit different environment and to form mutualistic relationships with different hosts. So it is assumable that different species of rhizobium can environmentally or genetically be modified to infect early non-leguminous crops or plants. Sesbania is such a legume [17, 22]. Detailed knowledge of the legume-rhizobia symbiosis at a molecular level would thus help in formulating strategies for developing potential rhizobia and non-legume-rhizobia symbiosis. This will have profound implication in the agricultural system of Bangladesh since the present use of nitrogen-rich fertilizers incurs high cost both on the economy and environment [18, 19].
Antibiotic resistance is an ancient and naturally occurring phenomenon widespread in the environment [33, 35]. The rhizosphere contains a mixture of metabolically active microbial populations that compete in this environment in relation to size, diversity and biochemical activity [33]. Production of antibiotics by some soil harbouring microorganisms mainly bacteria and fungi has been largely documented [33,36]. Such organisms include Streptomyces like Streptomyces coelicolor, Micromonospora purpurea and Streptomyces griseus which produce actinorhodin, gentamycin and streptomycin, respectively [33, 37]. Keeping these in mind the present study has been designed to understand the diversity of the strains and an attempt has been made to develop a simple, rapid, and cost-effective identification using antibiotics since the diversity and the controversial classification scheme of the strains pose a major problem [20, 21, 22, 34, 35]. Most of the rhizobial species harbour plasmids that vary in size and number. The nodulation (nodABC) genes, the regulatory (nodD) gene and the nitrogen fixing (nif/fix) genes are located on large (usually ≥ 100 kb) symbiotic plasmids (pSym) [31, 38]. Plasmid instability in rhizobia where genome rearrangements result in a loss or gain of symbiotic performance has been shown in a number of studies [31, 39]. A number of workers found that treatment of rhizobia with plasmid curing agents like acridine orange, acriflavine, UV etc. decreased the symbiotic properties of these bacteria, suggesting a link between the nitrogen fixation, nodulation and plasmids [31, 40]. The objective of this study was to explore the exciting avenues of research on causes (i.e. mutation, loss of plasmids) of rhizobial diversity. The further studies and experiments regarding this project can be determining the effects of physical and chemical mutagens on rhizobia followed by determination of plasmid transfer capabilities of the strain to other bacteria, which is of great agricultural significance, since plasmid transfer rhizobia in other related and non-related bacteria is regarded as one of the prime sources of strain diversity [31]. And with successful experiments of plasmid transfer between different bacterial species, we can achieve our goal of producing more nitrogen-rich foods that we consume every day [20- 22, 23-25, 31].
The objective of this study was to give a comparative account on the diversified nature of the strains collected from different legumes especially the Sesbania strain with the following specific approaches by isolation, purification, and characterization of the strains obtained from different legumes. Also, the observation of the rate of extracellular polysaccharide and lipopolysaccharide production by the strains along with analysis of antibiotic susceptibility pattern for strain identification and observing the plasmid profile of the strains.
MATERIALS AND METHODS
Isolation of root nodule bacteria from various legume plants
Collection of nodules
Six different types of legume plants were collected from different rural parts of Bangladesh. The plant samples were carefully transported to the research laboratory following all necessary procedure to keep the plants alive. Fresh, healthy and big nodules were carefully selected from each plant for study. The selected nodules were brown, dark brown and greyish. The color and freshness of the nodules indicated that an active fixation was been established between the nodule bacteria and the legume plants.
Surface sterilization of the nodules
Collected healthy nodules were thoroughly washed under tap water and then severed from the root using a sharp and clean cutter. Intact, undamaged nodules were then immersed in 95% ethanol for 5-1 seconds to break the surface tension, and then those were transferred to a 3% solution of H2O2 (Sigma-Aldrich, Riedstrabe, Germany) and soaked for 2-3 minutes. Nodules were then rinsed with sterile distilled water five changes using sterile forceps for transferring.
Isolation of root nodule bacteria
The primary step of the isolation process was to crush the sterile nodules with a blunt tipped glass rod in a large drop of sterile water in a petri dish. One loopful of nodule suspension was then streaked on Yeast Mannitol Agar (YMA) plate (Sigma-Aldrich, Riedstrabe, Germany). The same procedure was followed for every nodule sample. The designation of the isolates obtained and their respective hosts are listed in table 1.
Culture maintenance and preservation
For long term preservation the isolates were sub-cultured on YMA slants. After 24 hours of growth at 30 ºC sterile paraffin oil was added on the media and then stored at 4 ºC. Subcultures from these stock cultures were performed when needed [29].
Table 1. List of isolates and their respective host under investigation.
Identification of the isolates
Morphological colony characteristics of the isolates
The colony characteristics (i.e. shape, size, color, opacity, elevation, edge, margin of the bacterial colony and their growth rate) were determined by observing the colonies on YMA plates [29].
Cultural and metabolic characteristics
Presumptive test
Strains of rhizobia can be identified observing their growth on different solid and liquid media. The size, shape color, texture of the colonies and their ability to alter the pH of the media are generally stable characteristics and useful to determine or defining strains. Although the final decision as to whether a culture is or is not rhizobia is quite divisive due to its diversified nature and broad host range, it generally depends on plant infection tests. Other shreds of evidence can contribute at least to a presumptive decision [29, 30].
Growth on glucose peptone agar
Glucose-peptone agar media (Hi-Media, Mumbai, India) were used to differentiate rhizobia, which usually shows little or no growth on the media without altering the pH of the media. Contaminants like Agrobacteria shows massive growth media with a distinct change in pH and color of the media [29, 30].
Congo red test
The purity of the rhizobial isolates was detected by adding Congo red (0.25 g/100 ml of EtOH; 10 ml stock/liter of YMA) (Merck KGaA, Darmstadt, Germany) in YMA media. Most rhizobia absorb the dye only weakly whereas contaminants including Agrobacteria, take it up strongly [29, 30].
Confirmatory tests
To confirm whether the isolates were rhizobia or not, they were inoculated in different media for different physiochemical tests and then incubated depending on their growth rate at 30 ºC. The compositions of the media for each physiochemical test are described in the appendix.
Catalase activity test
The presence of the enzyme catalase in the rhizobial isolates was examined suspending one loopful of the organism in a drop of 3% H2O2 on a glass slide. Production of bubbles indicated a positive result or vice-versa [29, 30].
Citrate utilization test
The ability of the isolates to utilize citrate was determined by the growth of Simmon’s Citrate Agar (SCA) (Hi-Media, Mumbai, India). A distinct colour from green to blue referred to a positive utilization of citrate by the isolates [29, 30].
Production of exopolysaccharide
The production of exopolysaccharide by each isolate was noted. The appearance of the colonies (i.e., gummy, watery, translucent to a thick dense consistency, milky creamy appearance, opacity, presence of dark centers etc.) was observed [29, 30].
Determination of the antibiogram profile of the test strains
The Resistance of the test strains to different antibiotics were determined ‘in vivo’ by using the standardized agar disk diffusion method more commonly known as ‘The Kirby-Bauer Method’ [4]. A suspension of the test strain was prepared by adjusting the turbidity of the broth in phosphate buffered saline by comparing with that of McFarland 0.5 solution. With the help of a sterile glass rod a Muller-Hilton Agar plate (Sigma-Aldrich, Riedstrabe, Germany) (pH 7.0) was spread uniformly with 1ml of the strain solution. Antibiotic discs (Tetracycline, Ciprofloxacin, Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid, and Ampicillin) were applied aseptically on the surface of the inoculated plates at appropriate spatial arrangements by means of a sterile needle. The plates were then incubated at 30 ºC for 48-60 h. after incubation, the plates were observed for the presence of zones of inhibition and when present the diameters were measured in millimeters. The zone diameters for an individual antimicrobial agent were translated into susceptible, intermediate and resistant categories by referring to an interpreting Table (5) [7].
Screening of Plasmids and Plasmid Size
The modified method of Birnboim and Doly (1979)
This method for plasmid extraction was carried out according to the modified method of Birnboim and Doly, 1979 [5]. The steps are in chronological order: Fresh rhizobial culture (1 ml) was taken in Eppendorf tubes spun for 5 minutes at 13,000 rpm in a microcentrifuge. Then the supernatant was aspirated and the cells were re-suspended on solution 1 (500 µl) by re-centrifugation. The supernatant was removed and the pellet was completely re-suspended in (250 µl) of lysis buffer (L7). Then the tubes were kept at room temperature for 15 minutes.
After that, 250 µl of solution 3 was mixed gently with the sample 1 and 2, and precipitation buffer (N4) were mixed with sample 3 and 4. Then tubes were ice incubated for 30 minutes. Then the mixture was centrifuged for 5 minutes at 13,000 rpm to pellet the precipitated chromosomal DNA. The clean supernatants were transferred to fresh sterile Eppendorf tubes. Ice cold (-20 ºC) 95% ethanol (Sigma-Aldrich, Riedstrabe, Germany) (1 ml) were mixed with the sample, then kept or ice for 30 minutes, to precipitate the DNA before centrifugation for 5 minutes at 13,000 rpm. Then the supernatant was removed and the pellet was washed with 70% alcohol and dried and finally dry DNA was re-suspended in 50 µl of TE buffer.
The trough, with the agarose gel was placed in a tank. The gel was submerged with the TBE buffer. 25 µl of the extracted plasmid was mixed with 5 µl gel loading buffer (blue juice) and then poured in the wells of the agarose gel. The top of the tank was closed and connected to the power source. Electrophoresis was carried out at 110 volts for 1.5 hours, until the dye reached the end of the gel [27].
At the end of electrophoresis, the gel was replaced in a tray where 300 ml distilled water and 20 µl ethidium bromide solution was poured previously. The tray was then placed on shaker to shake for half an hour at 35 rpm. Then the gel was distained in distilled water and observed on a UV transilluminator [27].
The size or molecular weight of the plasmid was determined using a marker DNA of known molecular size, which was a one kb extension ladder. The distance traveled by the test strain plasmid was compared to the distance travelled by the marker DNA, and from this comparison, the size of the test plasmid was estimated [27].
Agarose gel electrophoresis (Agarose framing)
Agarose gel was prepared by dissolving 1% agarose and 1% sodium dodecyl sulphate in 40 ml Tris Borate Electrophoresis (TBE) buffer (Hi-Media, Mumbai, India) by boiling it and then allowing it to cool to about 50 ºC. The melted gel was then poured in the thoroughly cleaned trough, which was assembled with a well former or comb containing 12 teeth. The comb made the slots into which the samples were loaded. The gel was then allowed to solidify at room temperature, after which the comb was slowly and carefully removed [27].
RESULTS
Identification and characterization
Colony characteristics of the isolates
The growth rate of Pisum sativum/ Green Pea, Vigna mungo/ Urad bean and Lens culinaris/ Tan Lentils on YMA media is 48 hours whereas Sesbania aculeata/ Dhaincha was less than 48 hours and the growth rate of Phaseolus vulgaris/ Common Bean and Arachis hypogea/ Peanut have remained 48-72 hours (Figure 1). The circular milky white dense colonies with the low amount of LPS production has been observed in Pisum sativum/ Green Pea legumes and the growth rate of the isolate was same as the strain infective on Pisum spp. The Profuse amount of LPS production was observed in both Sesbania aculeata/ Dhaincha and Vigna mungo/ Urad bean but their colony characteristics are different such as circular, convex translucent colonies, smooth and glossy and circular convex, milky white colonies respectively. Interestingly, the growth rate of Sesbania aculeata/ Dhaincha was faster than other Sesbania strains whereas the Vigna mungo/Urad bean showed faster growth rate. On the other hand, the moderate amount of LPS/EPS production of Phaseolus vulgaris/Common Bean was observed with the isolate behaved accordingly as the strain infective on Phaseolus and circular, convex translucent colonies with a dark center that tends to merge whereas the circular, convex, entire colonies with profuse amount of LPS/EPS production was monitored in Lens culinaris/ Tan Lentils. But, circular, raised, convex opaque colonies with produce low amount of gum was observed in Arachis hypogea/ Peanut and the growth rate is faster than general Arachis spp. and produces less gum (Figure 1).
Cultural, Biochemical and metabolic characters
The presumptive test was performed for primary isolation of the test strains and to differentiate between the fast growing and slow growing strains. The results of the presumptive tests are shown below:
Growth on glucose peptone agar
Rhizobia do not grow or grow poorly on glucose peptone agar and do not alter the pH of the media but contaminants like Agrobacteria grow massively and change the pH of the media distinctly. Growth of the isolates on glucose peptone agar is summarized in table 2.
Table 2. Growth on glucose peptone agar.
Congo red test
Congo red test observed based on absorbance quality of the dye. In sample 1 and 2, both are shown as high absorbance of the dye. Contaminants can take up the dye strongly, the absorbance of the dye by Sample 1 and 2 is an unusual property. On the other hand, the low uptake of the dye Congo red is a usual characteristic feature of Rhizobium spp. in both sample 3, 4, 5 and 6 with poor absorbance of the dye (Figure 2).
Confirmatory tests
By performing different biochemical tests on different media, the purity of the test isolates was reconfirmed. The biochemical behaviour of the test strains is listed in table 3.
Table 3. Biochemical behavior of the test strains.
Production of exopolysaccharide
The production rates of exopolysaccharide of the isolates were observed as low, moderate and profuse with highly or less inactive on plants (Table 4).
Table 4. Production of exopolysaccharide.
Determination of antibiogram profile of the test strains
The zone of inhibition of S1 against Tetracycline, Ciprofloxacin, Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid and Ampicillin are 25 mm, 22 mm, 10 mm, 0 mm, 0 mm, 0 mm, 15 mm and 20 mm, respectively. It was showed sensitive against Tetracycline, Ciprofloxacin and Ampicillin whereas resistant against Bacitracin, Amoxicillin, Streptomycin and Erythromycin. Only Nalidixic acid was intermediate. The zone of inhibition of S2 against Tetracycline, Ciprofloxacin, Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid and Ampicillin are 09, 20, 0, 0, 08, 15, 26 and 0 mm respectively. It was showed sensitive against Ciprofloxacin and Nalidixic acid whereas resistant against Tetracycline, Bacitracin, Amoxicillin, Streptomycin and Ampicillin. Only Erythromycin was intermediate. The zone of inhibition of S3 against Tetracycline, Ciprofloxacin, Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid and Ampicillin are 32, 34, 0, 0, 0, 10, 0 and 0 mm respectively. It was showed sensitive against Tetracycline and Ciprofloxacin, whereas resistant against Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid and Ampicillin. The zone of inhibition of S4 against Tetracycline, Ciprofloxacin, Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid and Ampicillin are 24, 19, 13, 0, 15, 19, 30 and 09 mm, respectively. It was showed sensitive against Tetracycline, Ciprofloxacin, Streptomycin and Nalidixic acid whereas resistant against Amoxicillin and Ampicillin. Bacitracin and Erythromycin were intermediate. The zone of inhibition of S5 against Tetracycline, Ciprofloxacin, Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid and Ampicillin are 22, 20, 08, 0, 30, 14, 15 and 09 mm respectively. It was showed sensitive against Tetracycline, Ciprofloxacin and Streptomycin whereas resistant against Bacitracin, Amoxicillin and Ampicillin. Erythromycin and Nalidixic acid are intermediate. The zone of inhibition of S6 against Tetracycline, Ciprofloxacin, Bacitracin, Amoxicillin, Streptomycin, Erythromycin, Nalidixic acid and Ampicillin are 14, 0, 0, 0, 18, 32, 0 and 0 mm respectively. It was showed sensitive against Streptomycin and Erythromycin whereas resistant against Bacitracin, Amoxicillin, Streptomycin and Erythromycin. Only Nalidixic acid was intermediate. Antibiogram profile of the test strains are noted in Figure 3 (zone diameter) and table 5 (antibiotic tolerance).
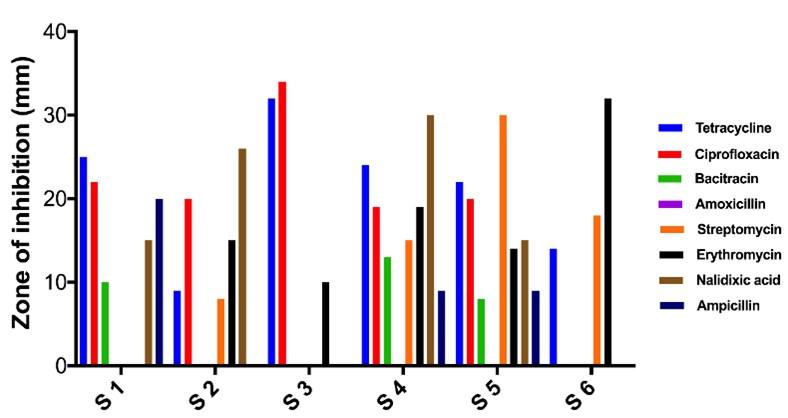
Table 5. Antibiogram profile (antibiotic tolerance).
Plasmid DNA profiling
Agarose gel electrophoresis of plasmid DNA preparations revealed that all the six (06) crystal producing isolates carried plasmids. The number of plasmid bands of the isolates ranges from 1 to 5. A common characteristic most of the isolates was the presence of a plasmid band above 564, 125 bp (Figure 4).
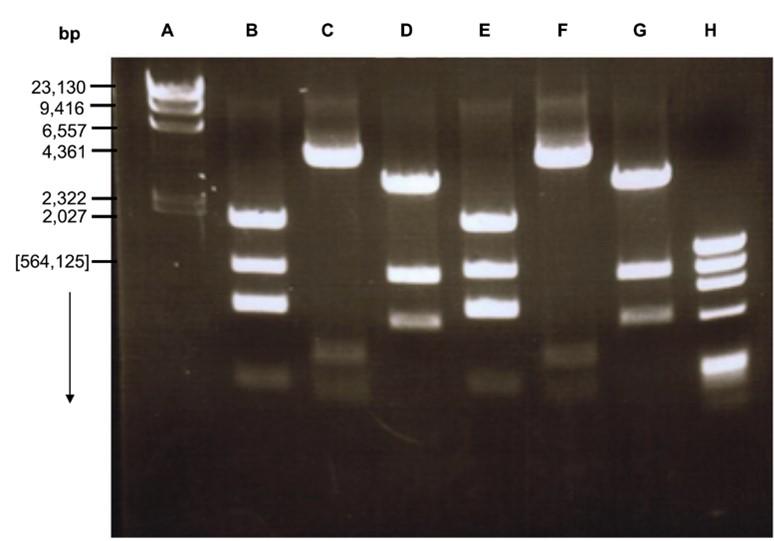
DISCUSSION
Endless curiosity and thirst for knowledge of scientists have revealed the mystery of the relationship between plants and bacteria to such a point that made a further study on this field a must. And one of the reasons for this is that, the mutualistic relationship or the symbiosis between plants and bacteria is a fact that is keeping plants and thus animals and human being alive [23-25, 27, 29, 31, 33, 40]. They are supplying the most necessary nitrogen to the planet. And today the vision has also been to use this miraculous relationship for human betterment. In third world countries like Bangladesh, where malnutrition is the most dangerous phenomena that keeping the population backward, further study on this field may reveal a way to remedy. The present study is expected to reveal the diversity of some very native rhizobial strains in Bangladesh, to some extent. Regarding their diversity, these strains have been investigated very poorly. But it can be hope for Bangladesh to get remedy from malnutrition of its peoples if we can reveal the mystery of symbiosis finally and use the knowledge for our purpose [23-25, 27, 29, 31, 33, 40].
The preliminary step of this experiment was to isolate and purify the rhizobial isolates collected from a variety of legumes (Table 1) from different ecological sites of Bangladesh, as the huge diversity of legumes is paralleled by a large diversity of rhizobial microsymbionts [6, 23-25, 27, 29, 31, 33, 40]. The test strains were identified and classified based on their cultural properties, biochemistry etc. the strains of rhizobia were found to be diverse in several properties.
The growth rate of the strains and their behaviour on different media were noted. The Sesbania (S 2), Vigna (S 3) and Arachis (S 6) strain exhibited rapid growth rate on various media, usually 48-72 hours. Normally these groups are sited in slow-growing groups in Rhizobia [7, 27, 29, 31, 33, 40] which needs ≥ 96 hours for growth. Therefore the tentative allocation of these strains in the slow-growing Bradyrhizobium [7, 27, 29, 31, 33, 40] group is seemingly unacceptable according to t the present study.
The Congo red, Yeast Mannitol Agar (YMA), and Glucose Peptone Agar (GPA) were employed to make presumptive decisions on the recognition and classification of the test strains. All the strains, except the Pisum (S 1) and Sesbania (S 2) strains showed poor absorbance of the dye Congo red. These facts give further evidence for the purity of rhizobial isolates [8, 31]. Poor growth on GPA can be explained as such that, rhizobia do not prefer peptone as the source of nitrogen, vitamins or growth factors of amino acids.
Absence of enzymes that cause breakdown and non-utilization of citrate serves as convenient diagnostic criteria for the identification of Rhizobia [9, 30, 31, 33, 40] investigated with 79 strains and reported that citrate utilization is a property exclusive to S. meliloti. In the experiment, no of the test strains showed the presence of enzymes that breakdown citrate.
The catalase activity test results of the sample strains differed from that of the previously studied strains. All the test strains posed catalase-negative, which is an unnatural phenomenon.
The production rate of EPS/LPS of rhizobia is an important fact that takes part in infecting plants. Also, there is an important relation between growth rate and EPS/LPS production of rhizobia. The growth rate is proportional to nitrogen fixation and EPS/LPS production determines the ability of plant infection or nodule formation of a strain. So in agronomical view, strains with high growth rate and high LPS/EPS production seek more importance [30, 31]. In this experiment, it was observed that The Sesbania strain had a faster growth rate with an ability to produce a profuse amount of LPS/EPS. Which was an unusual characteristic for the strain. The result indicated that this strain might be able to infect a host and fix nitrogen faster comparing with other strains. In this experiment, the strains Pisum and Arachis produced a low amount of LPS/EPS compared to its high growth rate and that is an unusual phenomenon for them [7, 31]. This indicated that these two strains might be able to fix nitrogen faster but will infect the host slowly. Like Sesbania, the strain Vigna also produced a high amount of EPS/LPS with a faster growth rate. Which proved its possible ability to infect a host and fix nitrogen faster. Lens, the strain was usual as its general growth rate but produced a high amount of EPS/LPS which indicated its possible ability to infect a host faster. About Phaseolus strain, it could be concluded that this strain might not be able to infect a host or fix nitrogen faster compared to other strains as its growth rate was normal and EPS/LPS producing ability was moderate. Finally, in agronomical view, it could be concluded that the Sesbania and Vigna strains are most important in the case of infecting hosts and fixing nitrogen.
The organism Rhizobia is strictly related to plants. There is no evidence in past that rhizobia have somehow interacted with animal or human immune system. Past studies [10, 31] recommended that antibiotic susceptibility/resistance pattern of rhizobia being useful as an identification tool, as this method is rather cost-effective and rapid. In this experiment, antibiogram profiling has done for a special purpose. Due to lack of time, the plant infection test could not be performed. This antibiogram profile of the test strains can be useful to retain our strains after plant infection test, as overlapping host range is a common phenomenon in symbiosis [31, 33]. In the present study, all the strains showed almost similar susceptibility/resistant pattern against the antibiotics used. More or less all the strains were resistant to bacitracin, Ampicillin and Amoxicillin which is natural for Rhizobium.
The process nitrogen fixation is a credit that goes only to the Sym plasmid. This plasmid is harbored by the organisms that can establish a symbiotic relationship with a legume. The modified method of Birnboim and Doly (1979) [5, 27, 31, 40] was applied for the extraction of the large rhizobial plasmids. It is expected that the Sym plasmids are usually ≥100 kb in size. A review on the S. meliloti genome reveals that the Sym B plasmid of the strain is extremely large, which is 1168 kb in size [11, 27, 40]. These plasmids pose a problem during extraction because they are prone to breakage. The resultant larger linear fragments are much more difficult to resolve by gel electrophoresis. Therefore, lysis of the cells with sodium dodecyl sulphate (SDS) in the agarose gel is more suitable for extracting large plasmids. In this experiment 1 kb extension ladder was used to determine plasmid size. But after the gel electrophoresis, no band was observed in the UV transilluminator. The plasmids of the strains might be lost or cured during the experiment. Or the organisms might have lost their Sym plasmids during the experiment as in experiment procedures the organisms were never exposed to such an environment where they might need that special plasmid. This kind of plasmid instability in rhizobia has been reported in several cases [12, 31].
CONCLUSIONS
The rhizobial strains present internal genomic dynamics that are continually generating subpopulations of similar but not identical organisms. Because of ever-increasing agronomic importance of rhizobia, it is indeed necessary that the causes and genetic basis of diversity of the strains and speciation of the genus be clarified. The presentation dealt with some of these interesting themes. The salient features of the present comparative study can be delineated as (i) comparison of different growth rates, biochemical and metabolic characteristics of the isolates with that of previously studied strains revealed that the strains are very much diverse, (ii) antibiogram of the wild type strains was more or less stable, and (iii) number of plasmid DNA band was observed under UV transilluminator after gel electrophoresis.
In conclusion, the diversity of the rhizobial strains is high; this may be due the gain of plasmids through transformation, conjugation events, presence of mutagens or even by spontaneous gene rearrangements. The diversity of the strains thus may be a consequence of the high rate of mutation, especially in their extrachromosomal DNA. In the end, this can be concluded that the antibiotic susceptibility/resistance pattern can form a basis for a marker in selecting these strains in any future work.
FUTURE PROSPECTS
In spite of the large diversity of the root nodulating bacteria, recent findings open new prospective and this will make rhizobial biology even more fascinating. The future work comprises of (i) to compare the genetic diversity of the strains for diversity analysis, (ii) study of the effect of different mutagens on the strains, (iii) transformation of different cells with the mutant strains, (iv) plant infection test for complete clarification of rhizobial species, (v) establishing protocols, like genomic fingerprinting, that provide high taxonomic resolution for rapid genotypic characterization of large collections of rhizobial isolates, (vi) plant infection tests with the wild type and mutant strains to observe and compare their nodulation and nitrogen-fixing trait, (vii) to construct a strain having desired genetic trait to increase nitrogen-fixing capability as compared to wild type strains, and (viii) finally, to construct strains capable of infecting rice and wheat crop plants and establish a symbiotic relationship that increases the nitrogen content in the crops and reduces the need for nitrogenous fertilizers.
ACKNOWLEDGEMENT
We thank Stamford University Bangladesh, Dhaka-1217, Bangladesh for providing us the facilities to carry out the experiments. However, the authors received no specific funding for this work.
AUTHOR CONTRIBUTIONS
This work was carried out in collaboration between all authors. All the authors have accepted responsibility for the entire content of this submitted manuscript and approved the submission. Authors AS, ATK, FYS and TBE performed experiments. AS, ATK, FYS, JM, MD, and TBE conceived the study and designed the experimental procedures. AS and TBE designed and planned the studies, supervised the experiments. TBE also acted for all correspondences. AS and TBE participated in the manuscript draft and has thoroughly checked and revised the manuscript for necessary changes in format, grammar and English standard. All authors read and approved the final version of the manuscript. All authors read and approved the manuscript.
CONFLICTS OF INTEREST
All authors declare no conflict of interest.
References
- [1]Wilde SA, Valley JW, Peck WH, Graham CM. Evidence from detrital zircons for the existence of continental crust and oceans on the Earth 4.4 Gyr ago. Nature 2001; 409 (6817): 175–8.
- [2]Adjei MB, Quesenberry KH, Chambliss CG. Nitrogen Fixation and Inoculation of Forage Legumes. IFAS Extension. SS-AGR-56, Institute of Food and Agricultural Sciences, University of Florida 2006.
- [3]Provorov NA, Vorobyov NI. Population genetics of Rhizobia: construction and analysis of an “infection and release” model. J. Theor. Biol. 2000; 205: 105- 199.
- [4]Barry AL, Thorsberry C. Susceptibility testing: diffusion disk procedure. In: Manual of Clinical Microbiology, 3rd edn., Novick, R.(ed). VCH publishing Co., New York 1985.
- [5]Birnboim HC, Doly J. A rapid procedure for screening recombinant alkaline plasmid DNA. Nucl. Acid Res. 1979; 7: 1513-1523.
- [6]Esperanza MR and Jesils CM. Rhizobium Phylogenies and Bacterial Genetic Diversity. Critical Rev. Plant Sci. 1996; 15(2): 113-140.
- [7]Bergey’s Manual of Systematic Bacteriology, 9th edition, 1984.
- [8]P. Somasegaran; R. Woolfenden; J. Halliday. Suitability of oven‐dried root nodules for Rhizobium strain identification by immunofluorescence and agglutination. J. Appl. Bacteriol. 1983.
- [9]Som Prasad Paudyal and Vimal NP Gupta. Bio-chemical characterization of rhizobia isolated from root nodules of Velvet bean (Mucuna pruriens L.). Our Nature 2017; 15 (1-2): 7-12.
- [10]Somasegaran and H. J. Hoben. Handbook for Rhizobia: Methods in legume-Rhizobium technology 1984.
- [11]Galibert F, Finan TM, Long SR, Puhler A, Capela D. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 2001; 27: 668-672.
- [12]Marta Laranjo, Ana Alexandre, Solange Oliveira. Legume growth-promoting rhizobia: An overview on the Mesorhizobium genus. Microbiol. Res. 2004; 169: 2-17.
- [13]Laranjoa M, Alexandrea A, Oliveira S. Legume growth-promoting rhizobia: An overview on the Mesorhizobium genus. Microbiol. Res. 2014; 169: 2–17.
- [14]Wdowiak-Wro´bel S; Marek-Kozaczuk M; Kalita M, Karas´ M; Wo´jcik M, Małek W. Diversity and plant growth promoting properties of rhizobia isolated from root nodules of Ononis arvensis. Antonie van Leeuwenhoek 2017; 110(8): 1087-1103.
- [15]Shamseldin, A., Abdelkhalek, A., and Sadowsky, M. J. Recent changes to the classification of symbiotic, nitrogen-fixing, legume-associating bacteria: a review. Symbiosis 2017; 71: 91–109.
- [16]Koskey G, Mburu SW, Kimiti JM, Ombori O, Maingi JM, Njeru EM. Genetic Characterization and Diversity of Rhizobium Isolated From Root Nodules of Mid-Altitude Climbing Bean (Phaseolus vulgaris L.) Varieties. Front. Microbiol. 2018 ,15;9:968.
- [17]Kuklinsky-Sobral, J., Araujo, W. L., Mendes, R., Geraldi, I. O., Pizzirani-Kleiner, A. A., and Azevedo, J. L. Isolation and characterization of soybean associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 2004; 6: 1244–1251.
- [18]Rahmani, H. A., Räsänen, L. A., Afshari, M., and Lindström, K. Genetic diversity and symbiotic effectiveness of rhizobia isolated from root nodules of Phaseolus vulgaris L. grown in soils of Iran. Appl. Soil Ecol. 2011; 48: 287–293. doi: 10.1016/j.apsoil.2011.04.010.
- [19]Anand, A., Jaiswal, S., Dhar, B., and Vaishampayan, A. Surviving and thriving in terms of symbiotic performance of antibiotic and phage-resistant mutants of Bradyrhizobium of soybean (Glycine max (L.) Merrill). Curr. Microbiol. 2012; 65: 390–397.
- [20]Benjelloun I, Thami Alami I, Douira A, Udupa SM. Phenotypic and Genotypic Diversity Among Symbiotic and Non-symbiotic Bacteria Present in Chickpea Nodules in Morocco. Front. Microbiol. 2019; 18: 10:1885.
- [21]Sinclair, M. J., and Eaglesham, A. R. J. Intrinsic antibiotic resistance in relation to colony morphology in three populations of West African cowpea rhizobia. Soil Biol. Biochem. 1984; 16: 247–251.
- [22]Rai, R., Dash, P. K., Mohapatra, T., and Singh, A. Phenotypic and molecular characterization of indigenous rhizobia nodulating chickpea in India. Indian J. Exp. Biol. 2012; 50: 340–350.
- [23]Chen W-M, Moulin L, Bontemps C, Vandamme P, Béna G, Boivin-Masson C. Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J. Bacteriol. 2003; 185: 7266–72.
- [24]Al-mujahidy, S. J., Hassan, M., and Rahman, M. Isolation and characterization of Rhizobium spp. and determination of their potency for growth factor production. Int. Res. J. Biotechnol.2013; 4: 117–123. doi: 10.14303/irjob. 2013.029.
- [25]Dai, J., Liu, X., and Wang, Y. Genetic diversity and phylogeny of rhizobia isolated from Caragana microphylla growing in desert soil in Ningxia, China. Genet. Mol. Res. 2012; 11: 2683–2693.
- [26]Boakye EY, Lawson IY, Danso SKA, Offei SK. Characterization and diversity of rhizobia nodulating selected tree legumes in Ghana. Symbiosis 2016; 69: 89–99. doi:10.1007/ s13199-016-0383-1.
- [27]Berrada, H., Nouioui, I., Houssaini, M., Ghachtouli, N., Gtari, M., and Benbrahim, K. Phenotypic and genotypic characterizations of rhizobia isolated from root nodules of multiple legume species native of Fez, Morocco. Afr. J. Microbiol. Res. 2012; 6: 5314-5324.
- [28]Gnat S, Małek W, Olen´ska E, Wdowiak-Wro´bel S, Kalita M, Łotocka B, Wo´jcik M. Phylogeny of symbiotic genes and the symbiotic properties of rhizobia specific to Astragalus glycyphyllos L. PLoS ONE. 2015; 10(10):e0141504. doi:10.1371/journal.pone.0141504.
- [29]Grönemeyer, J. L., Kulkarni, A., Berkelmann, D., Hurek, T., and Reinhold Hurek, B. Identification and characterization of rhizobia indigenous to the Okavango region in Sub-Saharan Africa. Appl. Environ. Microbiol. 2014; 4: 1–17. doi: 10.1128/AEM.02417-14.
- [30]Paudyal, S., & Gupta, V. N. Bio-chemical characterization of rhizobia isolated from root nodules of Velvet bean (Mucuna pruriens L.). Our Nature, 2017, 15(1-2): 7-12.
- [31]Nahar M, Mahal Z, Zahid HM., Zaman K, Jahan F, Rahman M.M and Noor R. Effects of plasmid curing on Rhizobium spp. Stamford J. Microbiol. 2012; 2(1): 34-37.
- [32]Ondieki DK, Nyaboga EN, Wagacha JM, Mwaura FB. Morphological and Genetic Diversity of Rhizobia Nodulating Cowpea (Vigna unguiculata L.) from Agricultural Soils of Lower Eastern Kenya. Int. J. Microbiol. 2017: 8684921.
- [33]Naamala J, Jaiswal SK, Dakora FD. Antibiotics Resistance in Rhizobium: Type, Process, Mechanism and Benefit for Agriculture. Curr. Microbiol. 2016; 72(6): 804-816.