Comparative molecular analysis of contemporary isolates of duck plague virus from haor areas of Bangladesh
Abstract
Duck plague (DP) is one of the most important viral diseases which affects the duck population across the globe including Bangladesh. The present work was conducted to detect DP virus (DPV) from haor areas using a molecular-based approach and compared with the contemporary isolate through molecular and phylogenetic analysis. For this purpose, 38 individual samples were collected from the Netrokona (n=20) district of the Mymensingh division and Kishoreganj (n=18) district of the Dhaka division. The identification of DVP was carried out by polymerase chain reaction (PCR) targeting DPV specific DNA polymerase genes followed by sequencing. PCR positive viral samples were used to propagate into 11-13 days old embryonated duck eggs through chorio-allantoic membrane (CAM) route for virus isolation. DPV were then propagated into duck embryo fibroblast (DEF) monolayer cell culture and confirmed by PCR. Among the 38 samples, 27 isolates were confirmed as DPV with the PCR amplicon size of 446 bp. Pathogenicity tests through the inoculation into day-old ducklings confirmed pathogenic strain. The PCR products of the isolated DPV specific DNA polymerase gene were sequenced commercially and submitted to GenBank (GenBank Accession No. KX768734.1). The sequence showed resemblance to isolates previously reported in India (GenBank Accession No. KX511893.1, KJ451479.1, KM012009.1), and China (GenBank Accession No. EF643559.1). Sequencing data also revealed nucleotide differences between Anatid herpes 1_BAU_DMH (previous report from our laboratory) and the present isolates. Further characterization, such as nucleotide and amino acid sequencing, would help to understand the strains along with its epidemiology.
INTRODUCTION
Duck plague (DP; synonym: duck viral enteritis), is the highly infectious and contagious disease of ducks, causing mortality of 60-70% [1] in Bangladesh. It is considered as one of the potential threats to commercially reared, domestic and wild waterfowl [2] throughout the world as it affects all age groups of ducks. DP is caused by duck herpesvirus 1 (anatid herpesvirus 1) under the genus Mardivirus of the family Herpesviridae and subfamily Alphaherpesvirinae [3]. Like other herpesviruses, this virus has a linear double-stranded DNA genome of 119×106 Daltons and comprised of approximately 158,091 base pairs [4]. 65 out of 67 genes are found as coding genes and three genes are unique to AHV-1 and the genome is made of unique long (UL), unique short (US), unique short internal repeat (IRS), and unique short terminal repeat (TRS) regions (UL-IRS-US-TRS in order) [4].
Direct or indirect contact with infected ducks or contaminated environment (that is contaminated with feces or oral and nasal secretions from an infected bird), respectively, can be the main mode of transmission of duck plague virus (DPV) [5]. Migratory waterfowls act as asymptomatic carriers of disease. In natural infection, incubation period varies from 3 to 7 days [6]. In most cases, the first notification is abrupt and consistently increasing flock mortality and infected ducks may die in good flesh though no symptoms are detectable. Affected birds show various general symptoms like other diseases, but prolapse of penis in dead mature male breeders and a marked reduction in egg production in female are remarkable, and mortality may reach up to 100% [7]. Diphtheroid plaques on the eyelids and the mucosae of the respiratory system and gastrointestinal system usually result in ophthalmic signs and refusal to water [6].
In the context of Bangladesh, the first report and confirmation of DPV had been made in the year 1982 [1], and later isolated and characterized by other investigators as well [8-10]. Hossain et al. [11] evaluated the immunogenicity of experimentally developed DPV vaccine from local isolates. DPV can be propagated and detected in duckling or adult duck [4], 9-12 days old embryonated duck eggs through chorio-allantoic membrane (CAM) route [9,12], avian fibroblast cell [13], kidney cell, liver cell etc. This virus can also be identified by passive haemagglutination (PHA) test [11,14]; neutralization test [15] or by polymerase chain reaction (PCR) with specific primer [10] and PCR is nearly 20 times more sensitive than tissue culture, and more reliable and accurate than traditional virus isolation and serologic identification methods used for detecting duck plague virus [16].
In Bangladesh, the total duck population is 54.016 million and ranked second in poultry, next to chicken [17]. Ducks are found throughout the country, especially in the north-east wetland ecosystem (haor) [18] and with more than 24% of the country’s total duck population found in this haor region [19]. In Netrokona and Kishoreganj districts, a higher number of duck populations is recorded than the other districts and provides self-employment for the landless and marginal small farmers in this region [18]. As DP is the most important disease of ducks, proper detection, isolation including molecular detection and analysis of the virus are of great importance. The molecular researches on duck plague virus are lag behind the other members of the herpesviridae family and even before this decade, mainly focused on epidemiology and prevention of the disease [20]. The genomic organization and nucleotide sequencing might be helpful for further study on duck plague virus [4]. Hence, UL30 gene (encoding DNA polymerase protein) based PCR detection, nucleotide sequencing and phylogenetic analysis was previously reported from our laboratory [10]. Subsequently, this present research work was undertaken as a follow up and comparative analysis to understand the strains and its epidemiology. Therefore, the present research was conducted for the isolation, identification, comparative molecular and phylogenetic analysis of DPV from haor areas in Bangladesh.
MATERIALS AND METHODS
Study area and period
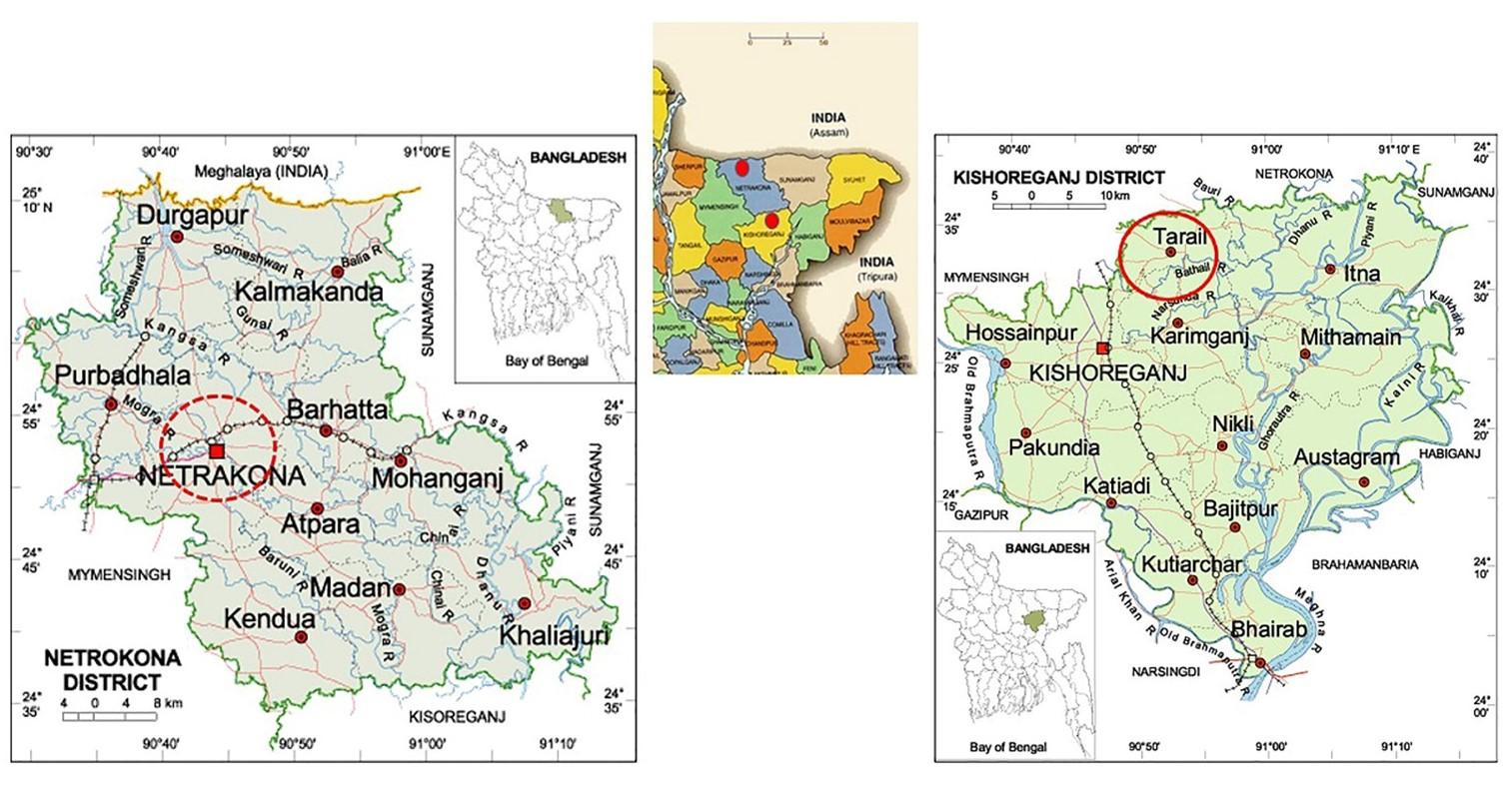
The experimental work was conducted at the Virology Laboratory, Department of Microbiology and Hygiene, BAU, Mymensingh, Bangladesh, from July 2015 to August 2016. A total of 38 randomly selected ducks (moribund and dead) from Sadar upazila of Netrokona district (n=20) of Mymensingh division and Tarail upazila (n=18) of Kishoreganj district of Dhaka division of Bangladesh. The map of the sampling locations is shown in the Figure 1.
Sampling
Different visceral organs, such as the esophagus, liver, intestine, and proventriculus, were collected aseptically during the post-mortem examination of 38 suspected DP affected ducks. The collected samples were kept separately in a sterile plastic container with proper labeling. Maintaining proper cool chain, the samples were then transported to the departmental Virology Laboratory, and few were processed immediately, and the rest were stored at ‑20°C until further analysis.
Preparation of inocula
The samples were processed by grinding, and PBS was added simultaneously to make a 10% suspension. Then centrifugation of the preliminary suspension was done at 4500 rpm for 10 min [2] and the supernatant fluid was collected and then treated with antibiotics (Gentamycin, 100μg/mL of suspension). For the sterility test, antibiotic-treated inocula were streaked separately with a sterile inoculating loop over the nutrient agar media and fresh blood agar media [11]. and the plate was incubated at 370C for overnight.
Propagation of virus in embryonated duck eggs
For this purpose, the prepared sterile inocula were inoculated into 11-13 days old embryonated duck eggs (EDE) through CAM route [2, 9], and at 6-8 days post-infection (PI), all dead and live EDEs was chilled overnight, and allantoic fluid and CAM were collected.
Propagation of the virus in duck embryo fibroblast cell culture
Duck embryo fibroblast cell culture was prepared according to the methods described by OIE manual [2] and Hossain et al [11]. Briefly, embryonated duck eggs (11-13 days old) were collected and soaked with 70% ethanol. The embryos were taken out and placed on a petri dish, followed by chopping of embryos excluding extremities and viscera. Chopped embryos were then taken into a conical flask where 0.25% trypsin was added for 100 mg tissues for cold trypsinization (40C for 18 hours), followed by the addition of 5% Minimum Essential Media (MEM) for minimization of effect of trypsin. Then the suspension was transferred into cell culture flasks and was incubated at 370C temperature and periodically observed for cell growth. Cell culture flasks containing 75-80% confluent growth of cells were taken for virus propagation. The remaining media of the cell culture flask was discarded. Cells were washed two times with PBS. About 200 μl virus inoculum was spread over monolayer duck embryo fibroblast cells. Then the cell culture flask was incubated at 370C for 60 min in an orbital shaker incubator to allow the virus to infect the cell. After incubation, 5% calf serum containing maintenance medium was added and incubated at 370C temperature. A cell culture flask was observed daily under an inverted microscope for the development of cytopathic effect (CPE).
Propagation of virus into ducklings
The duck plague virus was propagated into ducklings for pathogenicity test, according to the method described by Ahmad et al [10] and for this purpose, ethical approval was taken [Ethical Approval no. AWEEC/BAU/2017(09)]. For pathogenicity tests, about 500 µl (105.7 dELD50/mL) DPV suspension (CAM suspension) was inoculated in ducklings through the intramuscular route and observed for 6-8 days. It was expected that all the inoculated ducklings would be affected and would show clinical signs after 6 days PI.
DNA extraction, PCR and electrophoresis
DNA extraction kit Wizard® Genomic DNA Purification Kit (Promega, Madison, Wisconsin, United States) was used to extract DNA following the manufacturer’s instructions. The desired DNA segments of targeting the DNA polymerase gene of DPV (expected amplicon size of 446 bp) was amplified by PCR using the primers (Table 1) as described by Wu et al. [15]. The reaction mixture (total volume 25 µL) was comprised of nuclease-free water (7.5 µL), 2x PCR master mixture (12.5 µL; Promega, Madison, Wisconsin, United States), each forward and reverse primers (each 1 µL), and extracted DPV DNA template (3 µL). The thermal profile was set at initial denaturation at 94°C for 2 min; 35 cycles 94°C for 1 min, 56°C for 1 min, 72°C for 2 min, and, a final extension at 72°C for 7 min. For agarose gel electrophoresis of the PCR products, 2% agarose gel was used and PCR products (5 µL), including the DEV positive and native control, were mixed with 6X loading dye (Promega, USA; 1 µL) and loaded to the appropriate well. Agarose gel electrophoresis was accomplished following the method of Wu et al. [15].
Table 1. Primers for PCR.
Sequencing and analysis
For further molecular characterization, the PCR product of DNA polymerase gene (446-bp amplicon size) of DPV was used for partial sequencing. The PCR product was purified using Wizard® SV Gel and PCR Clean-Up system (Promega, Madison, Wisconsin, United States) according to their Quick protocol. Among the 27 positive isolates, two samples were sequenced, followed by analysis using BioEdit 7.0.5 Version [21] and ClustalX 2.0 [22] softwares. The finally obtained sequence was submitted to the GenBank. Then, the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/) was used to analyze the sequencing results, and comparison between the results of sequence analyses and a few other published sequences was made. For the construction of the Phylogenetic tree, the genome sequences of total 24 published sequences were aligned using CLC Sequence Viewer 8.0 software and finally, the phylogenetic analysis was accomplished through CLC Sequence Viewer 8.0 based on Neighbor-Joining method (Figure 7).
RESULTS
Isolation of duck plague virus
DPV was isolated from 27 samples out of 38, i.e., the overall isolation rate was 71.05% and district wise isolation rate were 80% (16/20) and 61.11% (11/18) in Netrokona and Kishoreganj district, respectively.
Propagation in embryonated duck eggs
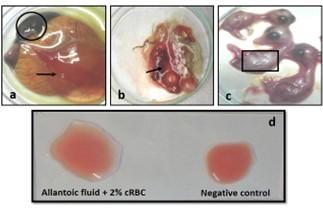
The allantoic fluid and the CAM were collected from the 11-13 days-old duck embryo which was inoculated with pure DPV suspension through CAM route. Embryo mortality in this study started from the 6 days PI and observed up to 8 days PI, and all the dead and live embryos were chilled at 4°C. Subcutaneous hemorrhages, thickened, and hemorrhagic CAM was found in dead embryos (Figure 2a, b, and c). Slide HA using the allantoic fluid revealed negative results (Figure 2d).
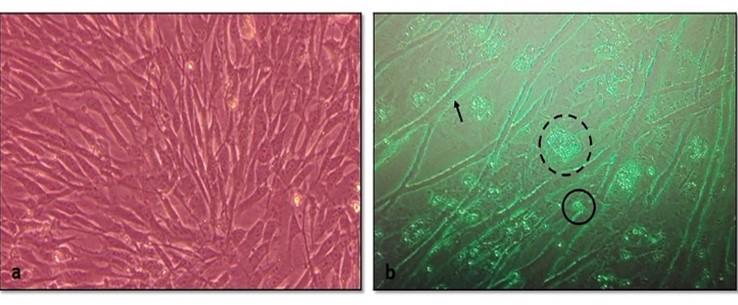
Cytopathic effects in duck embryo fibroblast cell culture
After the adaptation of the virus in fibroblast cells, CPE was observed as enlarged, rounded, clumped, degenerated, and necrosed of fibroblast cells and giant cell formation (Figure 3). Flasks with maximum CPE were frozen at -200C or -800C.
Pathogenicity test in day-old duckling
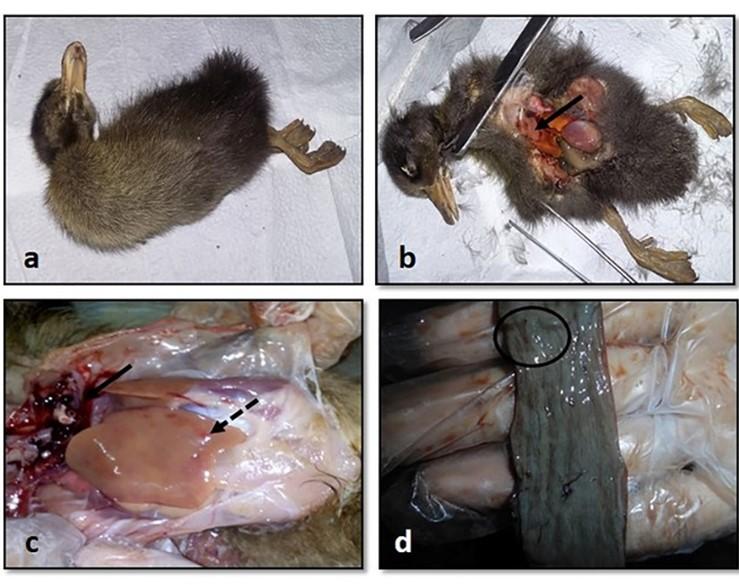
All the inoculated ducklings were affected within 6 days of post-infection showing clinical signs. Ducklings were found unable to stand with head down and drooped out-stretched wings and other symptoms, such as weakness, depression, off food, ataxia, and diarrhea were also found. Nervous sings began to rise as tremors of head, body, and neck, and finally death occurred (Figure 4a). Unclotted blood in the body cavities, pinpoint hemorrhages in the pale liver, and hemorrhagic annular band in the intestine were found on postmortem examination (Figure 4b, c, and d). For the re-isolation of the virus, visceral organs were processed and extracted for DNA and finally confirmed by PCR.
Detection of DPV by PCR
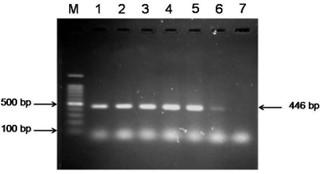
The extracted DNA samples from 38 field isolates were used to amplify DNA polymerase genes by PCR and 27 samples were found positive for DPV. All the PCR positive products showed the expected amplicon size of 446-bp (Figure 5).
Sequencing and phylogenetic analysis
The partially sequenced data were submitted to the GenBank (GenBank Accession No. KX768734.1) and the isolate was mentioned as BAU_DP_1T. Comparative analysis of DNA polymerase gene of DPV with previously reported isolates were performed and presented in Figure 6. The sequenced data were used to construct a phylogenetic tree (Figure 7). The tree revealed that the present isolates share the common ancestral origin with the isolates and strains found in Bangladesh, India, and China. The GenBank accession number and percent identities of the sequences of 24 isolates/strains were represented in the Table 2.

Table 2. List of isolates of DPV for homology study of fusion gene sequences.
DISCUSSION
Duck plague is considered the main obstacle in duck rearing and affects severely the large-scale duck farming in the haor and coastal areas of Bangladesh. The overall isolation rate (71.05%) of this study is in close agreement with the results reported by Hansen et al. [23] and Ahamed et al. [10]. In a previously published report from our laboratory, the overall prevalence was shown there as 66.67% [24] where the DP prevalence was 71.42% and 66.67% in Netrokona and Sunamganj district, respectively, whereas, in this study, that values were 80% and 61.11% in Netrokona and Kishoreganj district, respectively.
The cultural properties of the present isolates were observed both in embryonated duck eggs and cell culture system, and pathogenicity was indicated by inoculating in ducklings. The lesions found in this study (i.e., subcutaneous haemorrhage, thickened and hemorrhagic CAM) at 6 to 8 days PI, were consistent with the findings of Akter et al. [9], El-Samadony et al. [12] and Ahamed et al. [10]. As virus titre was highest in CAM, the CAM route is considered as the most sensitive way in the indicator host system for DPV propagation. In case of duck embryo fibroblast cell culture, the findings were supported by Nguyen et al. [25] and Aravind et al. [26] where the authors also observed similar CPE caused by DP virus in Duck embryo fibroblast cells. The pathogenicity test of the present isolates revealed that the isolates are pathogenic, as all the ducklings showed typical clinical signs within 6 days of post-infection. The results of the pathogenicity test is supported by the findings of El-Samadony et al. [12].
The PCR result of the present study was consistent and in agreement with the published reports of Wu et al. [15], Ahamed et al. [10], and OIE manual [2]. The use of the polymerase gene, for the PCR confirmation of DPV, is reported by various researchers from different countries of the world [27, 28].
Phylogenetic tree demonstrated that the isolate of duck plague virus (BAU_DP_1T) in this study was almost similar with the nucleic acid sequenced data found from GenBank. The GenBank Accession No. with KJ549663.1|: Anatidherpesvirus_1 stra CV, AF064639.1|:77-448 Anatid herpesvirus 1 DNA polymerase gene, EF643559.1|:635-932 Duck enteritis virus UL31 protein (UL31) gene, JQ647509.1|:59034-59405 Anatid herpesvirus 1 strain CHv and EU082088.2|:55521-55892 Duck enteritis virus strain VAC which has been reported as originated in China and was responsible for using duck plague in domestic ducks and other waterfowls. The phylogenetic tree also showed similarities with some other nucleic acid sequenced data from GenBank (Figure 7). We assume that the present isolate could be originated from the West Bengal and Kerala regions of India (KX511893.1: DEV/India/IVRI-2016; KM012009.1: isolate CDIO-duck01, Kerala, India 2015; KJ451479.1: DEV/Atpara/13, West Bengal, India 2014).
Comparative sequencing result with the previously published one [10], showed differences in few nucleotide base pairs (nucleotide position 374 to 379). Moreover, the previous authors reported only 378 bp among 446 bp PCR product. Therefore, further molecular analysis, such as nucleotide sequencing, amino acid sequencing etc. are of great significance.
CONCLUSION
Present research work revealed that the overall isolation rate was 71.05% (27 DPV positive isolates out of 38 suspected samples). PCR with DNA polymerase gene-specific primers and sequencing confirmed the isolates as DPV. The pathogenicity test also revealed that the field isolates were to be pathogenic. Nucleic acid sequencing of PCR products of 446-bp and phylogenetic tree showed that this isolate of duck plague virus (BAU_DP_1T) was highly similar with the isolates of DPV strain which were reported from India and China.
ACKNOWLEDGMENTS
None.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
AUTHOR CONTRIBUTIONS
MTK conducted the main research and both MTK and MBR was involved in conception and design of the experiment. MTRP and AK contributed to perform the experiment. TF helped in giving outbreaks information and sample collection. MAHS, MTR and KHMNHN were involved in sequence analysis. MTH and MPS contributed to prepare the manuscript and MBR finally made the approval of the version to be published. No funding was provided for this research work.
References
- [1]Sarker AJ. Duck plague in Bangladesh: Isolation and identification of the etiological agent. Indian Vet J. 1982; 59: 669-679.
- [2]OIE. Manual of diagnosis tests and vaccines for terrestrial animals; OIE Terrestrial Manual, 2018. Chapter 2.3.7. Duck Virus Enteritis.
- [3]ICTV. Virus taxonomy. EC 47. London, UK: 2015. [Accessed on 16-10-2016]. Available from: http://www.ictvonline.org/virustaxonomy.asp.
- [4]Li Y, Huang B, Ma X, Wu J, Li F, Ai W et al. Molecular characterization of the genome of duck enteritis virus. Virology. 2009; 391(2): 151-161.
- [5]Cheng AC, Wang MS, Liu F, Song Y, Yuan GP, Han XY et al. Distribution and Excretion of Duck Plague Virus Attenuated Cha Strain in Vaccinated Ducklings [J]. Chinese J Vet. 2005; 3: 002.
- [6]Maclachlan NJ, Dubovi EJ, Fenner F (Eds). Fenner’s Veterinary Virology. 4th ed. Amsterdam: Elsevier Academic Press. 2011, pp: 1914-2010.
- [7]Hoque MA, Skerratt LF, Cook AJ, Khan SA, Grace D, Alam MR et al. Factors limiting the health of semi-scavenging ducks in Bangladesh. Trop Anim Health Prod. 2011; 43: 441-50.
- [8]Islam MR, Khan MAHNA. An immunocytochemical study on the sequential tissue distribution of duck plague virus. Avian Pathol. 1995; 24(1): 189-194.
- [9]Akter S, Islam MA, Hossain MT, Begum MIA, Amin MM, Sadekuzzaman M. Characterization and pathogenicity of Duck Plague virus isolated from natural outbreaks in ducks of Bangladesh. Bang J Vet Med. 2004; 2: 107-111.
- [10]Ahamed MM, Hossain MT, Rahman M, Nazir KHMNH, Khan MFR, Parvej MS, et al. Molecular characterization of Duck Plague virus isolated from Bangladesh. J Adv Vet Anim Res. 2015; 2(3): 296-303.
- [11]Hossain MT, Islam MA, Amin MM, Islam MA. Comparative efficacy of the conventional and experimentally developed duck plague vaccine. Int J Poult Sci. 2005; 4: 369-371.
- [12]El-Samadony HA, Tantawy LA, Salama E, Khedr AA. Molecular Characterization of Circulating Duck Viral Enteritis in Egypt during 2012-2013. Br J Poult Sci. 2013; 2: 38-44.
- [13]Gao X, Jia R, Wang M, Zhu D, Chen S, Lin M et al. Construction and identification of a cDNA library for use in the yeast two-hybrid system from duck embryonic fibroblast cells post-infected with duck enteritis virus. Mol Biol Rep. 2014; 41: 467-475.
- [14]Das SC, Chowdhury SD, Khatun MA, Nishibori M, Isobe N, Yoshimura Y. Poultry production profile and expected future projection in Bangladesh. World Poultry Sci J. 2008; 64: 99-116.
- [15]Wu Y, Cheng A, Wang M, Zhang S, Zhu D, Jia AB et al. Serologic detection of duck enteritis virus using an indirect ELISA based on recombinant UL55 protein. Avian Dis. 2011; 55: 626-632.
- [16]Hansen WR, Brown SE, Nashold SW, Knudson DL. Identification of duck plague virus by polymerase chain reaction. Avian Dis. 1999; 43(1): 106-115.
- [17]DLS. Annual report on livestock. 2018. Department of Livestock Services, Ministry of Fisheries and Livestock, Farmgate, Dhaka, Bangladesh
- [18]Hamid MA. Duck genetic resources, their improvement and conservation in Bangladesh: a review. SAARC J Agric. 2019; 17(2): 31-42.
- [19]CEGIS, Center for Environmental and Geographic Information Services. 2012. Master Plan of Haor Areas. Volume II, Main Report. Bangladesh Haor and Wetland Development Board, Ministry of Water Resources, Government of the Peoples’ Republic of Bangladesh. Pp: 109.
- [20]Cai MS, Cheng AC, Wang MS, Chen WP, Zhang X, Zheng SX et al. Characterization of the duck plague virus UL35 gene. Intervirology. 2010; 53(6), 408-416.
- [21]Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series 1999; (Vol. 41, No. 41, pp. 95-98). [London]: Information Retrieval Ltd., c1979-c2000.
- [22]Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. Clustal W and Clustal X version 2.0. Bioinformatics, 2007; 23(21), 2947-2948.
- [23]Hansen WR, Nashold SW, Docherty DE, Brown SE, Knudson DL. Diagnosis of duck plague in waterfowl by polymerase chain reaction. Avian Dis. 2000; 44: 266-274.
- [24]Soma SS, Nazir KHMNH, Rahman MT, Rahman MM, Ara MS, Sultana R et al. Isolation and Molecular Detection of duck plague virus for the development of vaccine seed. Asian Australas J Biosci Biotechnol. 2018; 3(1): 78-85.
- [25]Nguyen TKD, Ho VH, Nguyen TTH, Do VD, Tran DT, Nind L et al. Adaptation of duck plague virus to chicken embryo fibroblast cell culture for vaccine production. Proceedings, No. 117. In: Meers J, Spradbrow PB, DhinTu Tran, editors. Control of Newcastle disease and duck plague in village poultry. Canberra, Australia: Australian Center for International Agricultural Research; 2004. p. 53-56.
- [26]Aravind S, Kamble NM, Gaikwad SS, Shukla SK, Dey S, Mohan CM. Adaptation and growth kinetics study of an Indian isolate of virulent duck enteritis virus in Vero cells. Microb Pathog. 2015; 78: 14-19.
- [27]Zhang D, Lai M, Cheng A, Wang M, Wu Y, Yang Q et al. Molecular characterization of the duck enteritis virus US10 protein. Virol J. 2017; 14(1): 183.
- [28]Mandal PS, Mukhopadhayay SK, Pradhan S, Mondal S, Jana C, Patra NC et al. Development of nested polymerase chain reaction-based diagnosis of duck enteritis virus and detection of DNA polymerase gene from non-descriptive duck breeds of West Bengal, India. Vet World, 2017; 10(3): 336.