Effect of chemical and physical mutagens on amylase producing potentiality of Aspergillus flavus NSH9
Abstract
Aspergillus flavus NSH9 is a potential source of α-amylase and glucoamylases. Between two of its glucoamylases, one can digest the raw starch granules, while both being pH and thermostable. The purpose of the study was to enhance the production of amylases (α-amylase and Glucoamylase) from A. flavus NSH9 by random mutagenesis using Ultraviolet (UV) irradiation and Ethidium bromide (EtBr) treatment. Glucoamylase and α-amylase activity were evaluated by 3, 5-dinitrosalicyclic acid (DNS) method and starch-iodine method, respectively. Mutated A. flavus NSH9 by EtBr treatment (10µg/ml) in PDA plate produced highest amount of both crude glucoamylases (GA) (1.47 ± 0.087 U/mL/min) and raw starch degrading glucoamylase (RSD-GA) (0.839 ± 0.036 U/mL/Hour). The highest value was 1.6 and 1.5 times higher compared to control for GA and RSD-GA, respectively. Here as, UV induced radiation produced about 1.3 and 1.4 times more GA and RSD-GA compared to control, respectively. The activity of α-amylase was about 1.7 times higher in the treatment group with 5µg/ml EtBr in sublethal condition than without the treatment group. In conclusion, both UV and EtBr treatment increased the amylases production from A. flavus NSH9. As the single mutation process of A. flavus NSH9 enhanced all three enzymes, the strains could be used for the commercial production of amylase.
INTRODUCTION
The amylases (α-amylases, β-amylases, and glucoamylases) are among the most valuable enzymes and are important for biotechnology, representing a class of industrial enzymes with approximately 30% of the world enzyme market [1-2]. Alpha amylase (1,4-α-D-glucan glucanohydrolase, E.C. 3.2.1.1) is a family of endo amylases that randomly cleaves the α-1,4 linkages throughout of the starch particles, generating glucose, maltose, dextrin, and oligosaccharides [3]. Whereas, glucoamylase (1,4-α-D-glucan glucohydrolase, E.C. 3.2.1.3, GA) is an exo-acting enzyme that produces β-D-glucose from the nonreducing ends of raw or soluble starches and associated polysaccharide chains by hydrolyzing α-1, 4 linkages [4]. Both amylases have a considerable role in starch processing in the food industries, for instance, for the glucose and fructose syrup derived from liquefied starch with both amylase and glucoamylase action [5-6]. Amylase is also used in different food processing, beverage, various fermented foods, pharmaceuticals, textile, and bioethanol industries [6-10]. Though, the sources of amylases are many such as plants, animals, bacteria and fungi, yet amylases of fungal origin are generally recognized as safe [9]. That is also the reason, for a significant percentage of fungi are continuously screened for α-amylase or glucoamylase production with properties that are better suited to specific industrial applications [11-13]. A. flavus has previously been recorded to be an active amylase enzyme producer [13-14]. We have previously purified and sequences one noble α-amylase gene; and two nobles thermostable glucoamylase were also sequences and expressed from A. favus NSH9 [15- 17]. One of its glucoamylase had starch binding domain (SBD) at C terminus and can degrade the raw starch granules [17].
Microorganisms can be easily controlled by means of genetic manipulation or other methods. These can be subject to strain development, mutations, genetic engineering, and other modifications that can increase the production of enzymes [18-19]. The techniques used for strain improvement can be applied either separately or in different combinations [20]. Random mutagenesis is widely used in the food industry for the classical strain development reasons [21]. The mutagens are of chemical or physical in nature. Chemical mutagenesis includes the use of Ethidium bromides (EtBr), ethyl methyl sulphonate (EMS), nitrous acid, and N-methyl N’nitro-N nitroguanidine (NTG) [22]. Physical mutagenesis encompasses UV rays, X-ray, and gamma rays. Among physical agents, UV treatment is commonly used in industries because it is very efficient and does not need any instruments [22]. The natural ability of microorganisms to create amylase has indeed been enhanced by mutational therapy, where both chemical and UV radiation mutagenesis have been shown to increase amylase activity [22-24]. Irradiation and chemical mutagens are mostly employed mutagens for mutations using random mutagenesis and some of them have considerably increased the characteristics of amylases in order to satisfy one or all of the 3 E’s i.e. Energy, Environment and Economy [25-27]. The purpose of the study was to improve the production of amylases (α-amylase and two glucoamylases) from A. flavus NSH9 through random mutagenesis by using UV radiation and EtBr at different concentration at sublethal condition.
MATERIALS AND METHODS
Culture media for enzyme production
A. flavus NSH9 has been used as an inoculum to produce amylase (α-amylase and two glucoamylases). So far as the enzyme production is concerned, active-growing fungal mycelium has been transferred from the potato dextrose agar ( PDA) plate to the minimum salt culture medium (MSM) containing (g / L): 4 g of yeast extract, 1 g (NH4)2SO4, 20 g raw sago starch, 3 g KH2PO4 and 0.5 g MgSO4.7H2O [18, 26]. The pH was set to pH 5.0 prior to autoclaving. Two pieces (approximately 5 mm in diameter of each cutted piece) of a 7-day-old PDA fungal crop were used for fermentation in 250 mL of a conical flask containing 50 mL of MSM medium containing 2% (w / v) of raw sago starch. The incubation was conducted at room temperature for 5 days on a rotary shaker at 150 rpm. [17, 28]. Details of sample preparation was discussed in the previous study [28].
Chemical and physical treatment for mutation
For chemical treatment in sublethal condition with EtBr at different concentration from 1µg/mL and 5µg/mL were added in the MNS culture media; and method is considered as treatment group one. The induction/fermentation procedure was also followed as discussed above.
Other chemical treatment with EtBr in PDA plate: In this method EtBr at concentration of 10µg/mL were added in potato dextrose agar (PDA) plate, and 100 µL of fungal spore suspension were spread onto the PDA plate. The process is considered as treatment group two. After that, the PDA plates were incubated for 7 days for growing mutated A. flavus NSH9. Two pieces of 7-day-old mutated fungal culture grown in PDA were then used for fermentation of 50 mL MSM medium. The incubation was carried out, as discussed above.
UV irradiation treatment was used as physical treatment for mutation and considered as treatment group three. Actively growing fungal mycelium was subjected to UV irradiation. First the fungal cultures grown actively for 5 days were suspended in 0.1M sodium acetate buffer (pH 5.0) and then were centrifuged for 10 min at 6000 rpm; this cell suspension was utilized for the mutation process. The mutation was induced by treatment with 10 mL of suspension which was aseptically pipetted into sterile flat-bottomed petri dishes. The sterile petri dishes containing the suspension were then exposed to UV light in a laminar airflow cabinet fitted with a germicidal lamp, according to Kumar et al. [29]. The samples were exposed for 30 min. The mutated fungal cell suspensions (150 µL/plate) were spread onto PDA medium for 7 days. Afterward, two pieces of 7-day-old mutated fungal culture grown in PDA were used for fermentation of 50 mL MSM medium. The incubation was carried out, as discussed above.
Glucoamylase assay
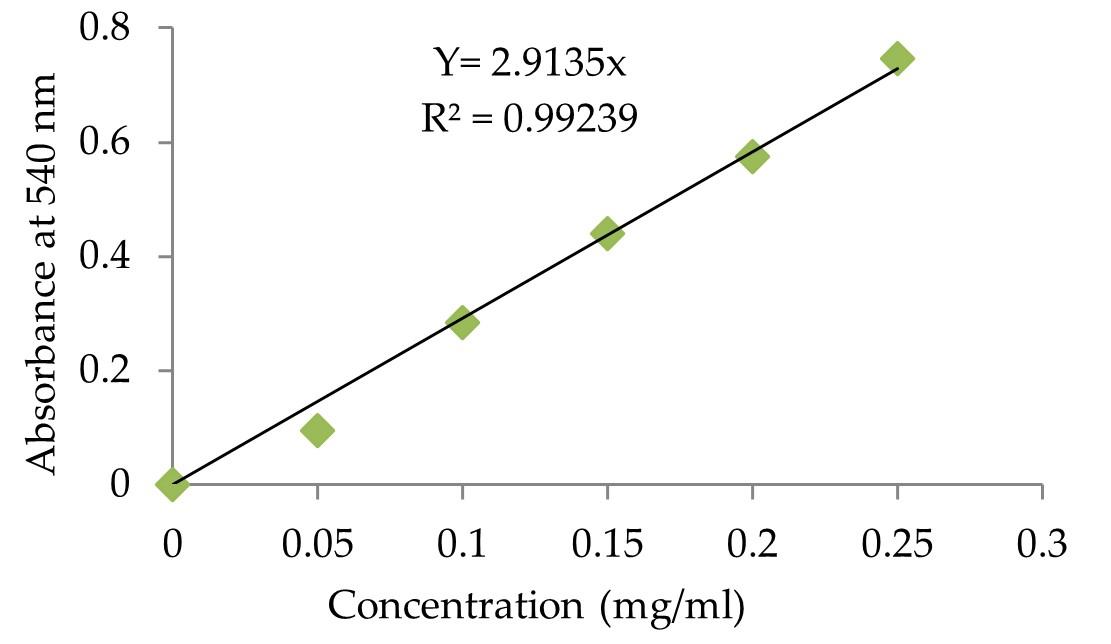
Glucoamylase activity was evaluated according to the technique used earlier by Karim et al. [15] and released glucose was measured using 3, 5-dinitrosalicyclic acid (DNS) method [30]. The absorbance measurements were performed at absorbance of 540 nm using a spectrophotometer. The enzyme activities were estimated using a calibration curve prepared with D-glucose as standard (Figure 1). One unit of glucoamylase activity is characterized as the amount of enzyme that releases 1 µ mole of glucose equivalent per minute from soluble starch under the test condition (at 55°C and pH 5.0) [15].
Raw starch degrading ability (RSDA) of glucoamylase
The degrading capacity of crude glucoamylase preparation to raw starch was determined by combining 0.5 mL of enzyme preparation with 0.5 mL of 1 percent (w / v) raw sago starch in 0.1 M sodium acetate buffer, pH 5.0. After 24 hours of incubation at 37 ° C with shaking at 150 rpm, the supernatant was collected for enzyme testing [28]. The reaction mixture was assayed as described in above GA assay using standard curve of glucose (Figure 1). One unit of the RSDA was defined as the amount of enzyme required to release one µ mole of glucose per hour under the assay conditions.
Alpha amylase estimation
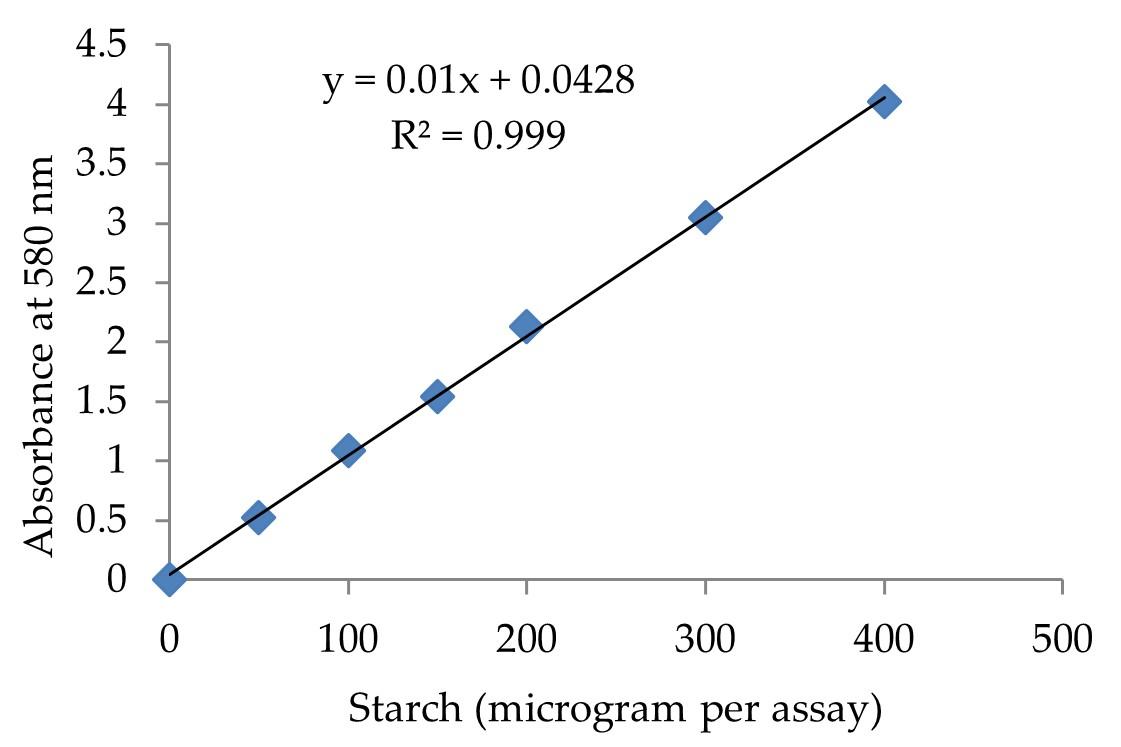
α-amylase activity has been calculated using starch-iodine method according to Xiao et al. [31] with minor modifications as discussed in previous study [16]. A standard curve of the starch-iodine complex was prepared using a different amount of starch in 400 μL samples containing 50–400 μg of starch (Figure 2), and the absorbance was measured at 580 nm. α-amylase activity unit (U) was defined in the starch-iodine assay as the disappearance in the assay reaction of an average of 1 mg of iodine binding starch material per min per mL.
RESULTS
Effect on crude glucoamylase
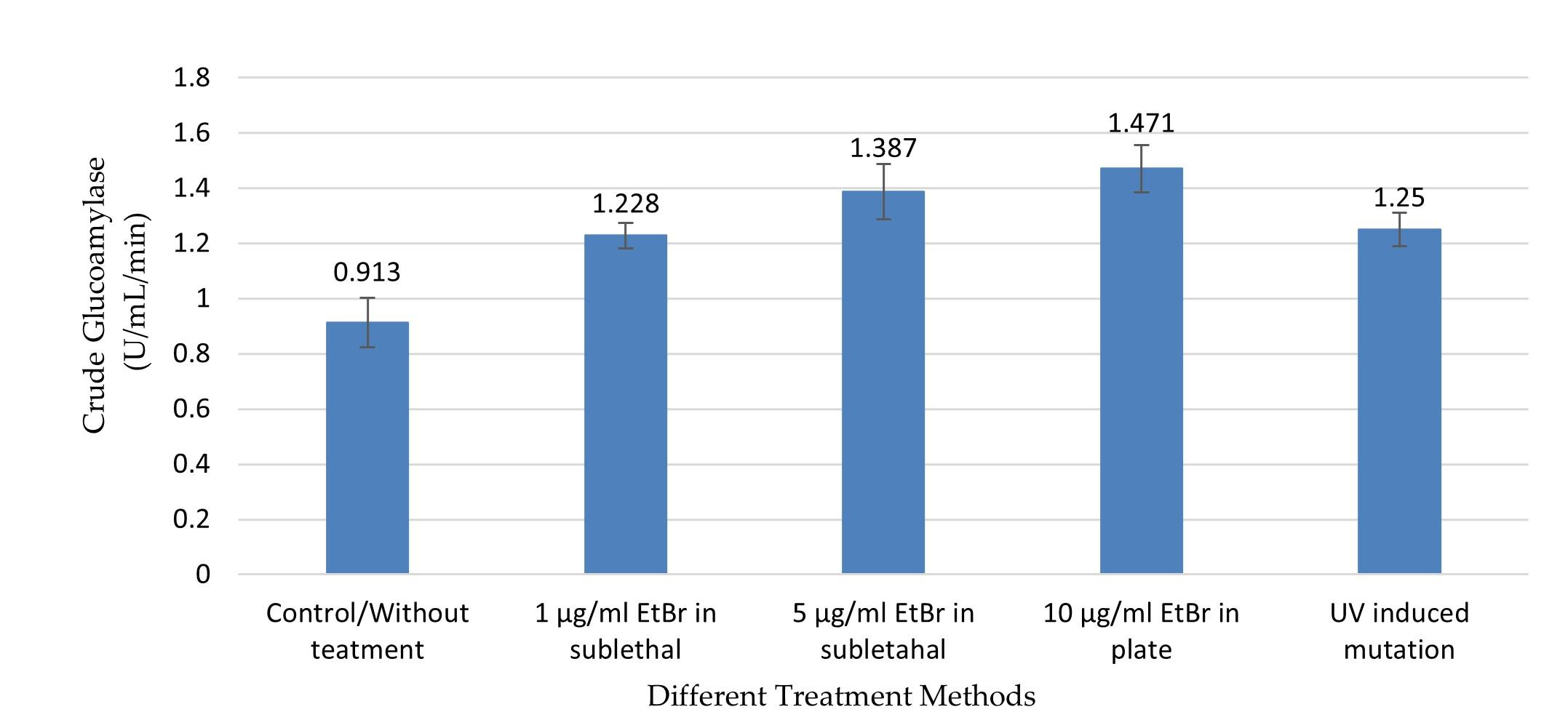
In this study, UV radiation and EtBr at different concentrations in sublethal condition and in PDA plate were used to improve the production of GA, RSD-GA, and α-amylase. The crude GA production was comparatively higher in different treatment groups compared to control (without treatment) group (0.913 ± 0.09), and the production was significantly (p ≤0.05) different between the groups as shown by one-way ANOVA (Figure 3). The highest GA activity (1.47 ± 0.087 U/mL/min) was observed in the culture media of the treatment group two with 10µg/ml EtBr in PDA plate method (Figure 3), and the value was about 1.6 times higher compared to without treatment group. This value was significantly higher from all groups (treatment and control) except the treatment group with 5 µg/ml EtBr (1.387 ± 0.101) in sublethal condition (p ≤0.05, by Tukey test).
Effect on raw starch degrading glucoamylase
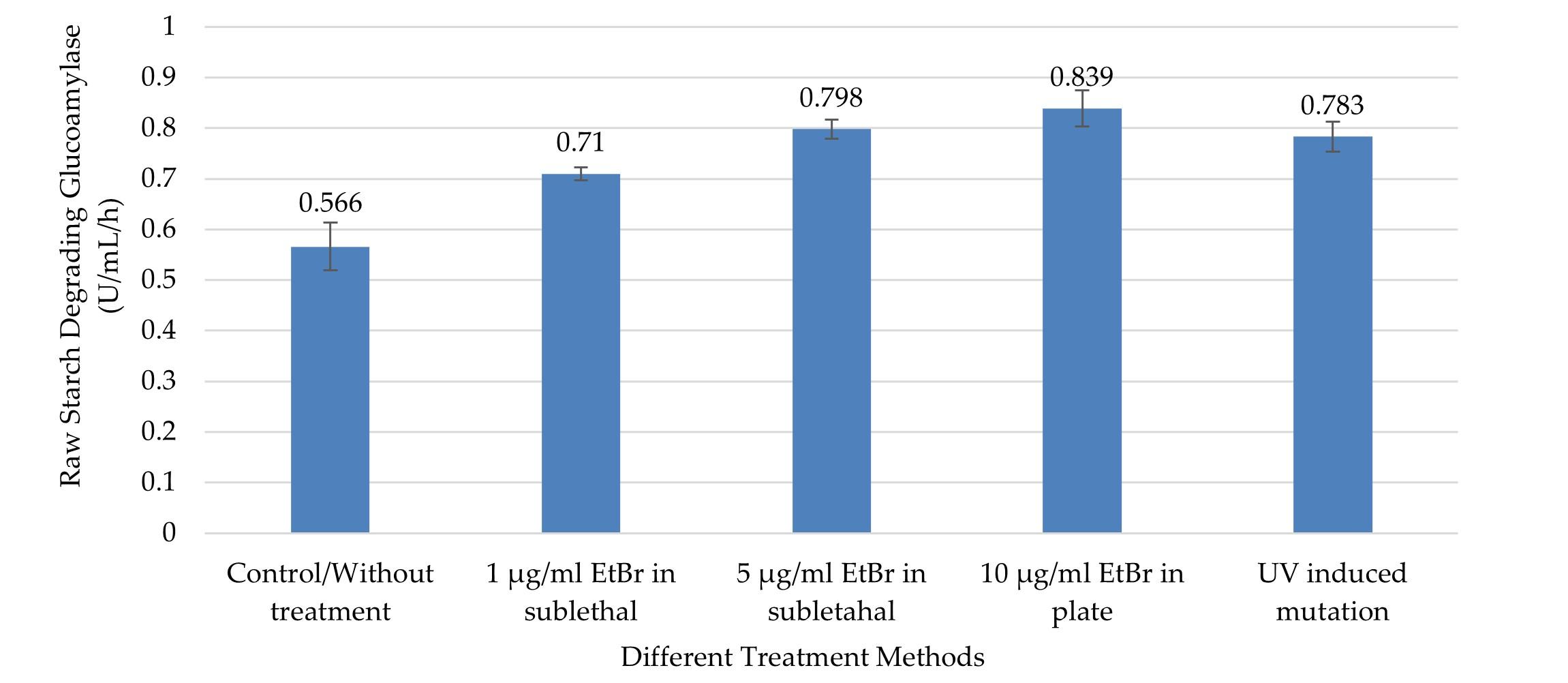
Like GA, RSD-GA production also gradually expanded along with increasing concentration of EtBr at sublethal state (Figure 4). As compared to control, all the treatment groups produced significantly higher RSD-GA (p ≤0.05). The maximum RSD-GA activity (0.839 ± 0.036 U/mL/Hour) was observed in the culture media of the treatment group two with 10µg/ml EtBr in PDA plate method (Figure 4). The highest value was about 1.5 times higher than control, which was remarkably close to treatment groups with UV and EtBr at 5µg/ml concentration (Figure 4). Meanwhile, UV induced radiation also produced about 1.4 times more RSD-GA as compared to control (Figure 4).
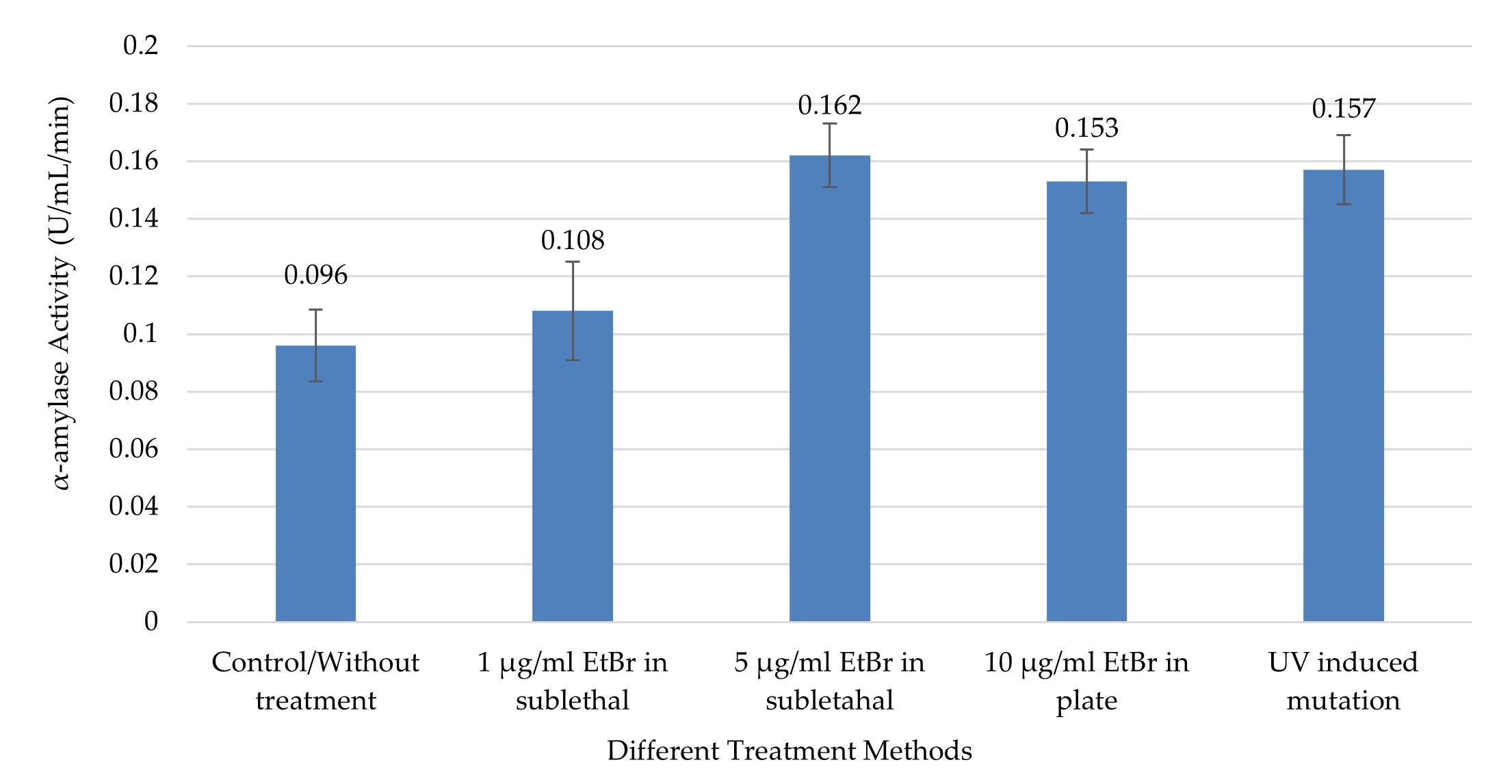
Effect on α-amylase
Like crude GA and RSD-GA, α-amylase production also differed significantly in various groups as found by one-way ANOVA (p ≤0.05). Compared to control, all the treatment groups produced significantly higher (p ≤0.05) α-amylase except the one with EtBr at low concentration (1µg/ml EtBr in sublethal condition).
The highest α-amylase activity (0.162 ± 0.011 U/mL/min) was observed with 5µg/ml EtBr in sublethal condition (Figure 5), and that was about 1.7 times higher than the control group. Meanwhile, UV treatment also produced about 1.63 times more α-amylase as compared to control (Figure 5). This highest value was significantly higher from only the control and treatment group with 1 µg/ml EtBr (0.108 ± 0.017) in sublethal condition (p ≤0.05, by Tukey test).
DISCUSSION
Previous studies demonstrated that A. flavus NSH9 is a potential candidate for α-amylase, GA, and RSD-GA [10, 15-17, 28]. So, it’s improvement through chemical and physical mutagens will be more industrial significance for amylases production [19]. This study shows the impact of chemical and physical mutagens on the ability of amylase production from A. flavus NSH9. Like previous studies, this study exhibited a significant increase in amylases (all three enzymes) production by mutagens EtBr and UV rays [25-26, 32-35]. The production of amylase (glucoamylase and amylase) by EtBr was higher compared to the UV radiation method in this study. A previous study observed that chemical mutagens (EtBr) are more capable of production glucoamylase from Aspergillus niger than physical mutagen (UV), which is similar to this study [32]. They also reported that combination of both the EtBr and EMS can give the best result for glucoamylase production [32]. About 1.5 times higher RSD-GA/RSDE activity was observed in this study with the EtBr treatment at 10µg/mL in PDA plate compared to wild strains, whereas a previous study recorded twofold higher RSDE from Aspergillus sp by combine treatments of γ-irradiation of Co60, UV and NTG [34]. The higher amount of α-amylase from the mutated A. flavus NSH9 by EtBr at 5μg/mL in sublethal condition was found in the study, which is comparable to the previous research in sublethal cellulose production [36]. Like the previous study, the chemically mutated strain produced a higher amount of α-amylase compare to UV mutated strains [26].
The research has few drawbacks, as the study used only one stage mutation technique, whereas the combination mutation method or the sequential mutation technique would have been better suited to the selection of potent mutants for the hyper-production of the desired enzyme. The research did not examine the nucleotide sequence of the intended mutated enzyme (amylases) to know the exact mutation point of the gene sequence. The study did not measure the toxic level of EtBr while using enzyme production at sublethal concentration.
CONCLUSION
The higher production of both glucoamylases was recorded from mutated A. flavus NSH9 by EtBr in the PDA plate. Whereas the highest α-amylase production was recorded from the same fungi by EtBr at a concentration of 5µg/ml in the culture media. Although chemical treatment appears to have been more effective in improving the production of all amylases by fungal strain testing, UV radiation also increased the production of all enzymes. As the single mutation process of A. flavus NSH9 enhanced all three amylase enzymes, the strains could be used in the industrial development of amylase, and thus could be potential sources of starch processing.
ACKNOWLEDGEMENT
None.
AUTHOR CONTRIBUTIONS
KMRK proposed the original idea and reviewed the scientific contents described in the manuscript. KMRK and AR performed the experiments and analyzed the data. Both KMRK and AR wrote the initial draft and reviewed the finial manuscripts. The authors received no financial support for the research, authorship, and publication of this manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Rodriguez VB, Alameda EJ, Gallegos J, Requena AR, Lopez A. Enzymatic Hydrolysis of Soluble Starch with an alpha amylase from Bacillus licheniformis. Biotechnol Progr. 2006; 22: 718–22.
- [2]van der Maarel, MJEC, van der Veen B, Uitdehaag, JCM, Leemhuis H. and Dijkhuizen L. Properties and applications of starch-converting enzymes of the alpha-amylase family. J Biotechnol. 2002; 94: 137–55.
- [3]Leman P, Goesaert H, and Delcour JA. Residual amylopectin structures of amylase treated wheat slurries reflect amylase mode of action. Food Hydrocoll. 2009; 23: 153-164.
- [4]Sauer J, Sigurskjold BW, Christensen U, Frandsen TP, Mirgorodskaya E, Harrison M, Roepstorff P, and Svensson B. Glucoamylase: structure/function relationships, and protein engineering. Biochim Biophys Acta. 2000; 1543: 275-293.
- [5]Nguyen QD, Rezessy-Szabo JM, Claeyssens M, Stals I and Hoschke A. Purification and characterization of amylolytic enzymes from thermophilic fungus Thermomyces lanuginosus strain ATCC 34626. Enzyme Microb Technol. 2002; 31: 345-52.
- [6]Abdalwahab SA, Ibrahim SA and Dawood ES. Culture condition for the production of glucoamylase enzyme by different isolates of Aspergillus spp. Int Food Res J. 2012; 19: 1261-66.
- [7]Pandey A, Nigam P, Soccol VT, Singh D and Mohan R. Advances in microbial enzymes. Biotechnol Appl Biochem. 2000; 31: 135-152.
- [8]Kumar P and Satyanarayana T. Microbial glucoamylases: characteristics and applications. Crit Rev Biotechnol. 2009; 29: 225-255.
- [9]Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B. Microbial α-amylases: a biotechnological perspective. Process Biochem. 2003; 38: 1599–1616
- [10]Karim KMR and Tasnim T. (2018). Fungal glucoamylase production and characterization: A review. Rioresearch Communications. 2018;4: 591-605.
- [11]Negi S and Banerjee R. Optimization of extraction and purification of glucoamylase produced by Aspergillus awamori in solid-state fermentation. Biotechnol Bioprocess Eng. 2009; 14: 60-66.
- [12]Sanghvi GV, Koyani RD and Rajput KS. Isolation, optimization, and partial purification of amylase from Chrysosporium asperatum by submerged fermentation. J Microbiol Biotechnol. 2011; 21: 470–176.
- [13]Fadahunsi IF and Garuba OE. Amylase production by Aspergillus flavus associated with the bio-deterioration of starch-based fermented foods. N Y Sci J. 2012; 5: 13-18.
- [14]El-Abyad MS, Fawzeya A, El-Sayed A and Hafez M. Effects of culture conditions on amylase production by some soil fungi. Zbl Mikrob. 1992; 147: 23-34.
- [15]Karim KM R, Husaini A, Hossain MA, Sing NN, Mohd Sinang F, Hussain MHM and Roslan HA. Heterologous, Expression, and Characterization of Thermostable Glucoamylase Derived from Aspergillus flavus NSH9 in Pichia pastoris. BioMed Res Int. 2016; 5962028.
- [16]Karim KMR, Husaini A, Sing NN, Mohd Sinang F, Roslan HA, Hussain H. Purification of an alpha amylase from Aspergillus flavus NSH9 and molecular characterization of its nucleotide gene sequence. 3 Biotech. 2018; 8. 204.
- [17]Karim KMR, Husaini A, Sing NN, Tasnim T, Sinang FM, Hussain H, Hossain MA and Roslan H. Characterization and Expression in Pichia pastoris of a Raw Starch Degrading Glucoamylase (GA2) derived from Aspergillus flavus NSH9. Protein Expr Purif. 2019;164: 105462.
- [18]Ajita S and Thirupathihalli PKM. α-Amylase Production and Applications: A Review. J Appl Environ Microbiol. 2014; 2: 166-175.
- [19]Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact. 2011; 10:32.
- [20]Nevalainen K. Strain improvement in filamentous fungi-an overview. Appl Microbiol Biotechnol. 2001; 1: 289–304.
- [21]Margolles A, Sanchez B. Selection of a Bifidobacterium animalis subsp. lactis strain with a decreased ability to produce acetic acid. Appl Environ Microbiol. 2012; 78: 3338–42.
- [22]Pathak SS, Sandhu SS, Rajak RC. 2015. Mutation studies on fungal glucoamylase: a review. Int J Pharm Biol Sci. 2015; 5: 297-308
- [23]EL-Bondkly AM and Keera AA. UV-and EMS-induced mutations affecting synthesis of alkaloids and lipase in Penicillium roquefortii. Arab J Biotechnol. 2007; 10: 241–248.
- [24]Vu VH, Pham TA, and Kim K. Improvement of fungal cellulase production by Mutation and optimization of solid state fermentation. Mycobiol. 2011; 39: 20–25.
- [25]Aleem B, Rashid MH, Zeb N. and et al. Random mutagenesis of super Koji (Aspergillus oryzae): improvement in production and thermal stability of α-amylases for maltose syrup production. BMC Microbiol. 2018; 18: 200.
- [26]Singh S, Singh S and Mangla J. Physical and Chemical Mutation for Enhanced Alpha-Amylase Production by Aspergillus fumigatus NTCC1222 under Solid State Fermentation Conditions Using Agri-Residue Waste. J Pharm Nutr Sci. 2016; 6: 22-26.
- [27]Singh S, Dutt D, Tyagi CH, Upadhyaya JS. Bio-conventional bleaching of wheat straw soda-AQ pulp with crude xylanases from SH-1 NTCC-1163 and SH-2 NTCC-1164 strains of Coprinellus disseminatus to mitigate AOX generation. New Biotechnol. 2010; 28: 47-57.
- [28]Karim KMR, Husaini A and Tasnim T. Production and Characterization of Crude Glucoamylase from Newely isolated Aspergillus flavus NSH9 in Liquid Culture. Am J Biochem Mol Biol. 2017; 7: 118-26.
- [29]Kumar GS, Chandra MS, Sumanth M, Vishnupriya A, Reddy BR, and Choi YL. Cellulolytic enzymes production from submerged fermentation of different substrates by newly isolated Bacillus spp. FME. K S J Appl Biol Chem. 2009; 52: 17-21.
- [30]Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem. 1959; 31: 426–428.
- [31]Xiao Z, Storms R and Tsang A. (2006). A quantitative starch-iodine method for measuring alpha-amylase and glucoamylase activities. Anal Biochem. 2006; 351: 146-48.
- [32]Raju EVN, Divakar G, Swetha C, Geetha J, Satish P. Strain improvement of Aspergillus niger for glucoamylase by physical and chemical mutagens. Int Res J Pharm App Sci. 2012; 2: 79-91.
- [33]Vu VH, Pham TM, Kim K. Improvement of a fungal strain by repeated and sequential mutagenesis and optimization of solid state fermentation for the hyperproduction of raw starch digesting enzyme. J Mirobiol Biotechnol. 2010; 20: 718-26.
- [34]Vu VH and Keun K. Hyper-production of raw starch-digesting enzyme by mutant fungal strain and optimization of solid byproducts. J Viet Env. 2012; 3: 66-70.
- [35]Oluwatoyin F, Olukunle and Oluwadamilola E, Ajayi. 2018. Screening Wild and Mutant Strains of Aspergillus flavus and Aspergillus niger Isolated from Plantain Stalks for Amylase Production. Jordan J Biol Sci. 2018; 11: 557 – 62.
- [36]Chand P, Aruna A, Maqsood AM and Rao LV. Novel mutation method for increased cellulase production. J. Appl. Microbiol. 2005; 98: 318-23.