Comparative polymer biodegradation efficiency of an isolated Acinetobacter sp. with Bravibacillus sp. and E. coli by resting cells
Abstract
At a concentration of 4 g/L, an enteric polymer is utilized to target drug release in the small intestine and causes considerable toxicity in cells. Our ecology and ecosystem are also harmed by their non-biodegradable qualities. We isolated and identified polymer-degrading bacteria from industrial effluent in this work. The isolated strain’s morphological, biochemical, and antibiotic sensitivities were also examined. The isolated strain was found to be gram-negative, round-shaped, and non-motile in morphological tests, while biochemical tests revealed it to be negative in starch agar and TSI but positive in methyl red, mannitol salt, simmon citrate, urea agar, and catalase test. The isolated strain was highly resistant to ciprofloxacin and vancomycin. The isolated bacterium was identified as Acinetobacter sp. by 16S rRNA sequencing. Additionally, Acinetobacter sp. strains of Escherichia coli and Brevibacillus sp. were used separately to observe the degradation of five synthesized non-biodegradable polymers (maleic acid propane-1,2 diol glycerol co-polyester, maleic acid phthalic acid propane-1,2 diol glycerol co-polyester, maleic acid phthalic acid butan-1,4 diol glycerol co-polyester, phthalic acid succinic acid propane-1,2 diol glycerol co-polyester, and phthalic acid succinic acid buten-1,4 diol glycerol co-polyester. The capacity of all three strains to degrade the above-mentioned polymers was greater than 75%. E. coli, for example, had a rapid disintegration rate but was responsible for human gastrointestinal and urinary tract infections. As a result, our isolated Acinetobacter sp. can be employed to degrade synthetic polymers.
INTRODUCTION
A polymer is a substance with a molecular structure that is made up of similar units and bonded together [1]. Polymers come in a variety of forms, including natural, synthetic, biodegradable, and non-biodegradable [2]. The enteric polymers have easy thermal processing, low price, and biodegradability. So, these enteric polymers can be utilized in the pharmaceutical and biomedical sectors [3]. Synthesis of newly designed polymers, the utilization of natural monomers, and chemical modification of current polymers are all viable options for achieving the stated objectives [4]. In today’s polymer science research, the development of biodegradable polymers plays a critical role [5]. Polylactic acid, polyglycolic acid, and other linear network polyesters with citric acid and polyglycerol co-polyester are biodegradable and have been used in medicine, and agriculture [5–7]. Polyester biodegradation occurs as a result of enzyme action and chemical breakdown in living organisms [8]. For linear network polyester biodegradation, a variety of microorganisms are used where bacteria including Enterobacter agglomerans, Serratia rubidaea, Pseudomonas aeruginosa, Staphylococcus epidermidis, Comamonas acidovorans, Corynebacterium sp. etc. [9–11]. The indigenous microorganisms are responsible for polymer biodegradable illumination [12]. Acinetobacter sp. was identified as a synthetic polymer degrading bacteria [13]. Antibiotic resistance in bacterial strains means that many drugs are ineffective against the bacteria. So, sensitivity analysis is determining, so that isolated bacteria are highly resistant to which specific drugs [14].
An enteric coating is a polymer barrier that is added to oral medications to prevent them from dissolving or disintegrating in the stomach [15]. This aids in either shielding drugs from stomach acidity, insulating the stomach from the drug’s negative effects, or releasing the drug after the stomach (typically in the upper intestine) [16]. Some drugs are sensitive to the acidic pH of the stomach and must be safeguarded from breakdown. Drug targeting can also be accomplished through the enteric coating (such as gastro-resistant drugs). Other drugs, such as anthelmintics, may require a high concentration in a specific area of the intestine [17]. The enteric coating can also be utilized as a research technique to measure drug absorption during trials [18]. The “delayed action” dosage form category includes enteric-coated medicines. From a pharmacological standpoint, the phrase “enteric coating” isn’t accurate, because gastric resistance can also be achieved by including enteric polymeric systems into the dosage form’s matrix. The most popular enteric-coated dosage forms include tablets, mini-tablets, pellets, and granules [19].
Recently, many advanced molecular culture-dependent techniques like library clones, LH-PCR (Length Heterogeneity Polymerase Chain Reaction), RISA (Rapid Interactive Structural Analysis), RT-Q-PCR (RT-Q-PCR Reverse Transcription Quantitative Real-Time PCR), RAPD (Random Amplified Polymorphic DNA), and RFLP (Restriction Fragment Length Polymorphism) have been developed and considered helpful tools for the isolation and identification of new bacterial strains. rDNA-dependent methods are rapid and reliable analyses of microbial cultures to identify microorganisms in comparison to traditional techniques. The 16s rRNA-specific molecular technique offers a better chance of identifying bacteria [20–22].
The polymer degradation is a major challenging issue for the environment due to their hazardous elements, which also pollute agricultural lands and reduce fertility. Some physiochemical recycling technologies are not environmentally friendly and cannot be used in all instances due to their high cost. So, in this study we isolated an ecofriend polymer degrading bacterial strain, and used morphological, biochemical, and molecular identification approaches to identify bacterial strain. Furthermore, synthetic polymer biodegradation capabilities were evaluated by resting cells using an estimated degradation rate (%) during the incubation period.
MATERIAL AND METHODS
Sample collection
Food industrial effluent was collected from Pran Agro Ltd. (latitude 24.838524°N and longitude 88.910172°E) Natore, Rajshahi, Bangladesh, and two other strains E. coli (Accession: NOLW03000003.1) and Brevibacillus sp. (Accession: MK517601.1) were collected from the Microbiology Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi, Bangladesh. Five different synthesized co-polyester samples such, maleic acid propane-1,2 diol glycerol co-polyester (P-1), maleic acid phthalic acid propane-1,2 diol glycerol co-polyester (P-2), maleic acid phthalic acid butan-1,4 diol glycerol co-polyester (P-3), phthalic acid succinic acid propane-1,2 diol glycerol co-polyester (P-4), and phthalic acid succinic acid buten-1,4 diol glycerol co-polyester (P-5) were collected from Hybrid Engineering Material Lab, Department of Applied Chemistry and Chemical Engineering, University of Rajshahi, Rajshahi, Bangladesh.
Bacterial isolation and morphological characterization
The strain was isolated by using the LB agar plate method [23]. Various serially diluted samples were placed into agar plates and incubated for 24 h at 37°C. Then the chosen colony was cultured several times by the steak plate method. Morphological characteristics were examined and recorded according to the method described by Cheesbrough, 1984 [25] and motility was measured according to Jarrell and McBride, 2008 [26].
Biochemical test
The biochemical characterization of isolated strain was done according to the following method reported by Paul et al., 2020 [23]. In biochemical characterization, Methyl Red, Catalase, MacConkey, Starch Agar, Mannitol Salt Agar, TSI (Triple Sugar Iron), Simmons Citrate Agar, and Urease tests were performed.
Antibiotic sensitivity test
Commercially available and frequently prescribed antibiotics such as Penicillin (10 units/disc), Amoxicillin (30 mcg/disc), Ciprofloxacin (5 mcg/disc), Erythromycin (15 mcg/disc), Gentamycin (10 mcg/disc), Vancomycin (30 mcg/disc), and Chloramphenicol (30 mcg/disc) were utilized in the antibiotic sensitivity test. This was accomplished by placing 109 CFU/mL of freshly cultured isolated bacterial strain on agar plates and inserting antibiotic disks into the plates. The plates were then incubated at 37°C for an overnight period. The zone was observed and quantified on a millimeter-scale after incubation.
Molecular identification
The genomic DNA was isolated using the method described by Cheng and Jiang, 2006, in which the 16S rRNA gene was amplified by PCR and sequenced for molecular identification of bacteria. For amplification, universal forward primer 27F – 5ˊ-AGAGTTTGATCCTGGCTCAG-3ˊ and reverse primer 1492R – 5ˊ- GGTTACCTTGTTACGACT-3ˊ for amplification [27] were used. The amplified 16S rRNA gene fragment was purified and sequenced using the Sanger sequencing method [28], using the same primer used for PCR amplification. 16S rRNA gene sequences were sequenced and aligned by comparing them to other sequences from the gene bank database using the Basic Local Alignment Search Tool (BLAST) available from the website (www.ncbi.nlm.nih.gov/Blast) to identify bacteria [29].
Polymer degradation by bacterial resting cells
For this, 100 mL of nutrient broth medium containing 1 mg/L of synthetic co-polymers samples (Dissolved 3 mg of each sample in 0.6 ml DMSO (Dimethyl Sulfoxide) to get the concentration of 5μg μL-1 with 5% DMSO) in 250 ml Erlenmeyer flask was applied for culturing and incubating in MS medium at 37°C for 72 h. Every 12h, 24h, 36h, 48h, and 72h, 10 ml of an aliquot of the culture broth was collected and centrifuged at 5000 rpm for 5 min at 4°C. The supernatant was taken and analyzed by UV-visible Spectrophotometer (Analytical Jena, Germany) at a wavelength of 440 nm. The degradation rate was calculated according to the following formula [30].
% of polymer degradation = × 100
Statistical analysis
For statistical analysis, this experiment was replicated three times for each biological sample. Duncan’s Multiple Range Test (DMRT) was an analysis of the significance of each group data at P≤ 0.05 label of significance at one-way ANOVA in SPSS Statistics 26 software. Graph Pad Prism 8.0.2.263 was used for all figures preparation.
RESULTS
Morphological and biochemical identification
Morphological characteristics of the isolated strain are shown in Table 1. The isolated strain was distinguished by its nearly round, gram-negative, yellowish, raised, and entire smooth appearance. Biochemical features are shown in Table 2, which indicated that the isolated strain was positive for Methyl Red, Catalase, MacConkey, Mannitol salt, and Simmons citrate tests and negative for Starch agar, and TSI tests.
Table 1. Morphological characteristics of isolated bacterial strain.
Table 2. Biochemical characterization of isolated bacterial stain.
Molecular identification
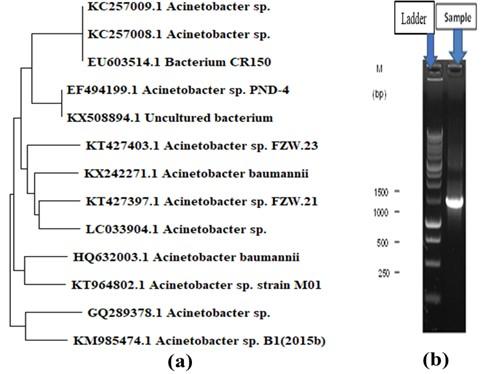
The isolated bacterial strain shared 95% of its DNA with Acinetobacter sp. and the constructed phylogenetic tree is shown in Figure 1a. Almost 1400 bp of targeted amp icons were amplified by PCR and are shown in Figure 1b.
Antibiotic sensitivity of isolate strain
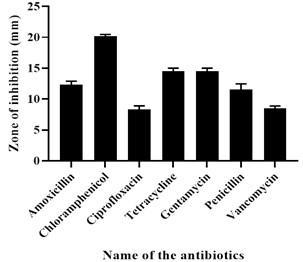
The result showed that the isolated strain was susceptible to Chloramphenicol, Vancomycin, Gentamycin, and Ciprofloxacin but intermediate resistant to Erythromycin and resistant to Penicillin and Amoxicillin. The result of the antibiotic sensitivity test is shown in Figure 2.
Synthesized polymer degradation
Various patterns for polymer degradation rate by growing cells of Acinetobacter sp., E. coli, and Brevibacillus sp. were observed, recorded, and detected and are shown in Figure (3-4) and Table 3. The degradation rate was measured by bacterial growing cells. In the case of maleic acid propane-1,2 diol glycerol co-polyester polymer degradation, E. coli, Brevibacillus sp., and Acinetobacter sp. showed 96.36 %, 95.97 %, and 97.02 % degradation respectively whereas in Maleic acid phthalic acid propane-1,2 diol glycerol co-polyester E. coli, Brevibacillus sp., and Acinetobacter sp. showed 94.09 %, 92.86 %, and 89.15 % percent degradation respectively. and in maleic acid phthalic acid butan-1,4 diol glycerol co-polyester, E. coli, Brevibacillus sp., and Acinetobacter sp. showed 87.86 %, 82.12 %, and 77.95 % degradation after 72 hours of incubation. In Phthalic acid succinic acid propane-1,2 diol glycerol co-polyester E. coli, Brevibacillus sp., and Acinetobacter sp. showed 93.47 %, 83.50 %, and 75.75 % degradation and in Phthalic acid succinic acid buten-1,4 diol glycerol co-polyester polyester E. coli, Brevibacillus sp., and Acinetobacter sp. were showed 78.91 %, 77.77 %, and 73.90 % degradation respectively. These results suggested that all three strains degraded five synthetic polymers and E. coli showed the highest level of degradation. Both Bravibacillus sp. and Acinetobacter sp. also demonstrated considerable polymer breakdown after 72 hours of incubation.
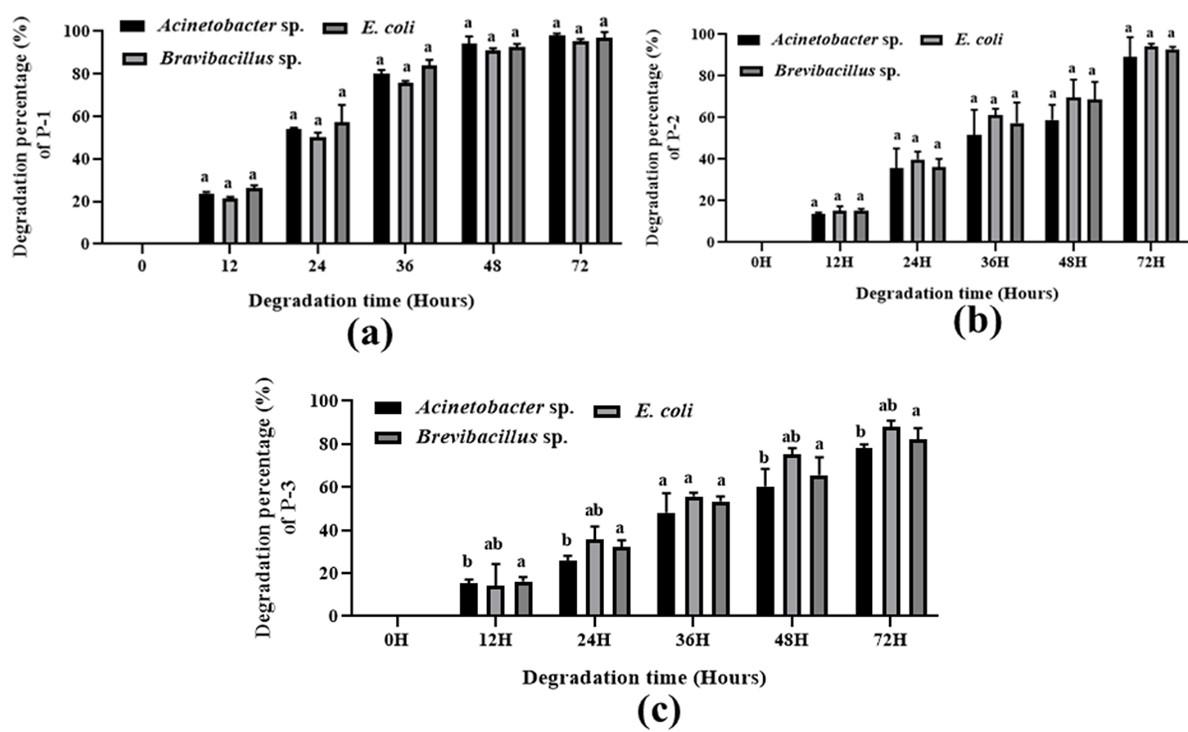
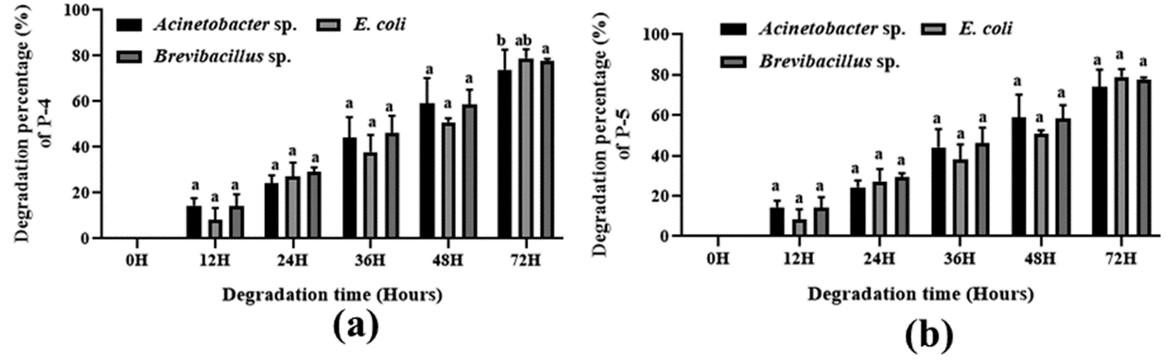
Table 3. Comparative polymer degradation percentage of isolated Acinetobacter sp., E. coli, and Brevibacillus sp. by bacterial growing cells after 72 hours of incubation.
DISCUSSION
Plastics are large-scale chemically generated long-chain polymers that have become an integral element of our society due to their low cost [32,33]. The rate of polymer deposition has accelerated dramatically in the last two decades, and it has also been imposed on the marine environment, destroying the marine ecosystem [33]. In the intestines of fish, birds, and marine mammals, it also creates obstructions. Moreover, entanglement with or ingestion of this trash has put hundreds of different species in jeopardy [34–36]. There are several physical and chemical ways of degrading polymers, but they are all quite expensive. As a result, there is a pressing need for low-cost, environmentally acceptable polymer breakdown processes. Bioremediation is a non-hazardous, cost-effective, and ecologically benign alternative approach for polymer breakdown. According to a survey on plastic garbage output in 60 major Indian cities, the country produces about 15,340 tons of plastic waste each day (Central Pollution Control Board (CPCB) New Delhi, India, 2013), which is tremendously hazardous to the environment.
In this investigation, we isolated a polymer-degrading bacterial strain from industrial effluent and performed morphological and biochemical characterization to confirm the isolate’s identity. Methyl Red, Catalase, MacConkey, Mannitol salt, and Simmons citrate tests were positive for our isolated strain, whereas Starch agar and TSI tests were negative. According to a recent study, Acinetobacter sp. strains isolated from hospital units [37] had nearly identical biochemical properties, including being negative in mannitol and sucrose, H2S (Hydrogen Sulfide) on TSI, nitrate reduction, and methyl red [37]. The isolate was susceptible to chloramphenicol, Vancomycin, Gentamycin, and Ciprofloxacin in an antibiotic sensitivity test, but intermediate resistance to erythromycin and resistant to penicillin and amoxicillin. According to a recent study, wastewater treatment plants are a rich source of antibiotic-resistant intestinal bacteria and genes that can be passed on to other bacteria in the environment [38].
Due to their high chemical inert complexity, only a tiny number of microbial organisms discovered on Earth are capable of digesting artificially manufactured polymers. Brevibacillus borstelensis, Rhodococcus rubber, Bacillus circulans, B brevies, B pumilus, B cereus, Staphylococcus epidermidis, Pseudomonas aeruginosa, Shewanella putrefaciens, and Nocardia asteroids were among the bacteria recently recovered from industrial wastewater [39–46]. The capacity of bacteria to produce biofilm on polymer surfaces enhances polymer breakdown, and our isolated strain may have this potential. Both E. coli and Brevibacillus sp. showed significant degradation on chosen synthetic polymers in our study, while isolated Acinetobacter sp. also showed significant degradation. The rate of Maleic acid propane-1,2 diol glycerol co-polyester polymer degradation was 97.02 %, maleic acid phthalic acid propane-1,2 diol glycerol co-polyester polymer degradation was 89.15 %, maleic acid phthalic acid butan-1,4 diol glycerol co-polyester polymer degradation was 77.95 %, phthalic acid succinic acid propane-1,2 diol glycerol co-polyester degradation was 75.75 % and phthalic acid succinic acid buten-1,4 diol glycerol co-polyester polyester degradation was 73.90 % by Acinetobacter sp.
Clostridium botulinum and Alcaligenes faecalis were previously identified as PCL degraders in a previous study [47]. Within 10 days, P. lilacinus D218 degraded the PCL by 10%, according to [48]. In PCL and PHB-containing media, P. lilacinus D218 produces PCL de-polymerases in addition to PHB de-polymerase. PCL de-polymerases were found to have the best activity at 30°C and pH levels between 3.5 and 4.5 [48]. [49] observed 92% degradation of 10 g PCL with particle size 125–250 m after anaerobic biodegradation at 55 °C with sludge (diluted 0.86% and undiluted 1.73%). In another study observed that the degradation of biodegradable bags was higher than polyethylene (PE) bags, with 100% degradation of the compostable material between 16 and 24 weeks [50]. Biofilm formation on the plastic bag surface after 15 days of exposure to the marine environment [51]. Microbial polymers degradation has recently been recorded as more economical and eco-friendlier than physiochemical methods. Bioremediation, especially using bacteria, is thus becoming an evolving and significant polymers treatment field.
CONCLUSIONS
Synthetic polymer production is expanding every day, causing pollution in the environment. The goal of this research is to find an ecofriendly bacterial strain that can biodegrade manmade polymers. Resting cells of this isolated Acinetobacter sp. displayed a substantial polymer degradation capacity. However, the study does not assess the cytotoxicity of the released components. It should be evaluated in the future, and specific enzymes from the strain should be identified for large-scale application.
ACKNOWLEDGMENT
We acknowledge Microbiology Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi, Rajshahi, Bangladesh, and Hybrid Engineering Material Lab, Department of Applied Chemistry and Chemical Engineering, University of Rajshahi, Rajshahi, Bangladesh
AUTHOR CONTRIBUTIONS
SA, GKP, and MSU conceived the idea and planned the research. SA and GKP also performed all experiments. GKP, SA, and SM prepared the manuscript. MSU supervised the research and MAS, and SZ revised the manuscript. All the authors read and approved the manuscript for publication. This research work receives no external funding.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Hoskins B, Robson R. Design and construction of a new class of scaffolding-like materials comprising infinite polymeric frameworks of 3D-linked molecular rods. A reappraisal of the zinc cyanide and cadmium cyanide structures and the synthesis and structure of the diamond-rela. J Am Chem. 1990; 112(4):1546–54.
- [2]Kluin OS, Van Der Mei HC, Busscher HJ, Neut D. Biodegradable vs non-biodegradable antibiotic delivery devices in the treatment of osteomyelitis. Expert Opin Drug Deliv. 2013; 10(3):341–51.
- [3]anke SSS, Gayakwad SG, Strom JG, D’Souza MJ. Oral delivery of low molecular weight heparin microspheres prepared using biodegradable polymer matrix system. J Microencapsul. 2009; 26(6):493–500.
- [4]Okada H. Nature and development of Cretaceous sedimentary basins in East Asia: a review. Geosci. J. 2000; 4(4): 271-282.
- [5]slam S, Haque M, Hossain S, Bakr A. Malic Acid Butane-1, 4-Diol-Glycerol Co-Polyester as an Enteric Coating Material. 2015; 3(6): 4–8.
- [6]Vroman I, Tighzert L. Biodegradable Polymers. Materials. 2009; 2(2): 307–44.
- [7]Chandra R, Rustgi R. Pergamon BIODEGRADABLE POLYMERS. Prog. Polym. Sci. 1998; 23(7):1273–335.
- [8]Rodriguez-galan A, Franco L, Puiggali J. Degradable Poly(ester amide)s for Biomedical Applications. Polymers. 2011; 3(1): 65–99.
- [9]Kay MJ, Morton LHG, Prince EL. Bacterial degradation of polyester polyurethane. Int Biodeterior. 1991;27(2): 205–22.
- [10]Kay MJ, McCabe RW, Morton LHG. Chemical and physical changes occurring in polyester polyurethane during biodegradation. Int Biodeterior Biodegrad. 1993;31(3):209–25.
- [11]Nair SS, Reddy H, Ganjewala D. Screening and Characterization of Biopolymers Polyhydroxybutyrate producing Bacteria. Adv Biotech. 2008;7:13–8.
- [12]Nair SS, Ganjewala D. Screening and characterization of Biopolymers Polyhydroxybutyrate producing bacteria. Adv. Biotechnol. 2016;7:13-17.
- [13]Penkhrue W, Khanongnuch C, Masaki K, Pathom-aree W, Punyodom W, Lumyong S. Isolation and screening of biopolymer-degrading microorganisms from northern Thailand. World J Microbiol Biotechnol. 2015;31(9):1431–42.
- [14]Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol. ASM.2009; 15: 55-63.
- [15]Lee PI. Polymers for controlled drug delivery. J Control Release. 1994;29:202–3.
- [16]Illum L, Bundgaard H. Sorption of drugs by plastic infusion bags. Int J Pharm. 1982;10:339–51.
- [17]Hansen BL, Hansen GN, Hagen C. Immunoreactive material resembling ovine prolactin in perikarya and nerve terminals of the rat hypothalamus. Cell Tissue Res. 1982;226(1):121–31.
- [18]Hong Wen KP. Oral Controlled Release Formulation Design and Drug Delivery. John Wiley Sons, Inc. 2010.
- [19]Hall GCN, Hong JJ, Zane NWS, Meyer OL. Culturally Competent Treatments for Asian Americans: The Relevance of Mindfulness and Acceptance-Based Psychotherapies. Clin Psychol Sci Pract. 2011;18(3):215–31.
- [20]Udgire M, Shah N, Jadhav M. Enrichment , Isolation and Identification of Hydrocarbon Degrading Bacteria. IntJCurrMicrobiolAppSci. 2015;4:708–13.
- [21]Teske A, Sigalevich P, Cohen Y, Muyzer G. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl Environ Microbiol. 1996;62(2):4210–5.
- [22]Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. Biotechniques. 1994;17(1):144-146.
- [23]Paul GK, Mahmud S, Naher K, Jabin T, Mahmud ML, Haque MN, et al. Isolation and characterization of bacteria from two soil samples and their effect on wheat (Triticum aestivum l.) growth promotion. J Adv Biotechnol Exp Ther. 2020;3(3):254–62.
- [24]Cheesbrough M. Medical laboratory manual for tropical countries. Trop Heal Technol UK. 1984;1:214–5.
- [25]Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6(6):466–76.
- [26]Cheng HR, Jiang N. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 2006;28:55–9.
- [27]Jeong Soon K, Gang Gweon L, Jong Seok P, Yong Hyun J, Hyo Sun K, Soo Bok K, et al. A novel multiplex PCR assay for rapid and simultaneous detection of five pathogenic bacteria: Escherichia coli O157:H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J Food Prot. 2007;70:70 (7) 1656-1662.
- [28]Koonin E V., Mushegian AR, Rudd KE. Sequencing and analysis of bacterial genomes. Curr Biol. 1996;6:404–16.
- [29]Stach JE., Bathe S, Clapp JP, Burns RG. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol Ecol. 2006;36:139–51.
- [30]Bhange VP, William SP, Sharma A, Gabhane J, Vaidya AN, Wate SR. Pretreatment of garden biomass using Fenton’s reagent: Influence of Fe2+and H2O2 concentrations on lignocellulose degradation. J Environ Heal Sci Eng. 2015;13.
- [31]Zheng Y, Yanful EK, Bassi AS. A review of plastic waste biodegradation. Crit Rev Biotechnol. 2005;25:243–50.
- [32]Alex S. New perspectives in plastic biodegradation. Curr Opin Biotechnol. 2011;22:422–6.
- [33]Jayasiri HB, Purushothaman CS, Vennila A. Quantitative analysis of plastic debris on recreational beaches in Mumbai, India. Mar Pollut Bull. 2013;77:107–12.
- [34]Teuten EL, Saquing JM, Knappe DRU, Barlaz MA, Jonsson S, Björn A, et al. Transport and release of chemicals from plastics to the environment and to wildlife. Philos Trans R Soc B Biol Sci. 2009;364:2027–45.
- [35]Secchi ER, Zarzur S. Plastic debris ingested by a Blainville ’ s beaked whale , Mesoplodon densirostris , washed ashore in Brazil. Aquat Mamm. 1999;25:21–4.
- [36]Reicher SD, Spears R, Postmes T. A Social Identity Model of Deindividuation Phenomena. Eur Rev Soc Psychol. 1995;6:161–98.
- [37]Filimon R, Taraşi I. Cultural and Biochemical Characteristics of Acinetobacter Spp . Strains Isolated From Hospital Units. J Prev Med. 2004;12:35–42.
- [38]Finley RL, Collignon P, Larsson DGJ, Mcewen SA, Li XZ, Gaze WH, et al. The scourge of antibiotic resistance: The important role of the environment. Clin Infect Dis. 2013;57:704–10.
- [39]Sudhakar M, Doble M, Murthy PS, Venkatesan R. Marine microbe-mediated biodegradation of low- and high-density polyethylenes. Int Biodeterior Biodegrad. 2008;61:203–13.
- [40]Bonhomme S, Cuer A, Delort AM, Lemaire J, Sancelme M, Scott G. Environmental biodegradation of polyethylene. Polym Degrad Stab. 2003;81:441–52.
- [41]Gilan I, Hadar Y, Sivan A. Colonization, biofilm formation and biodegradation of polyethylene by a strain of Rhodococcus ruber. Appl Microbiol Biotechnol. 2004;65:97–104.
- [42]Hadad D, Geresh S, Sivan A. Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J Appl Microbiol. 2005;98:1093–100.
- [43]Marchal R, Nicolau E, Ballaguet JP, Bertoncini F. Biodegradability of polyethylene glycol 400 by complex microfloras. Int Biodeterior Biodegrad. 2008;62:384–90.
- [44]Roy PK, Titus S, Surekha P, Tulsi E, Deshmukh C, Rajagopal C. Degradation of abiotically aged LDPE films containing pro-oxidant by bacterial consortium. Polym Degrad Stab. 2008;93:1917–22.
- [45]Watanabe T, Ohtake Y, Asabe H, Murakami N, Furukawa M. Biodegradability and degrading microbes of low-density polyethylene. J Appl Polym Sci. 2009;111:551–9.
- [46]Chatterjee S, Roy B, Roy D, Banerjee R. Enzyme-mediated biodegradation of heat treated commercial polyethylene by Staphylococcal species. Polym Degrad Stab. 2010;95:195–200.
- [47]Tokiwa Y, Calabia BP, Ugwu CU, Aiba S. Biodegradability of plastics. Int J Mol Sci. 2009;10:3722–42.
- [48]Oda T, Wals P, Osterburg HH, Johnson SA, Pasinetti GM, Morgan TE, et al. Clusterin (apoJ) alters the aggregation of amyloid β-peptide (Aβ1-42) and forms slowly sedimenting aβ complexes that cause oxidative stress. Exp Neurol. 1995;136:22–31.
- [49]Yagi H, Ninomiya F, Funabashi M, Kunioka M. Anaerobic biodegradation tests of poly(lactic acid) and polycaprolactone using new evaluation system for methane fermentation in anaerobic sludge. Polym Degrad Stab. 2009;94:1397–404.
- [50]Urbanek AK, Rymowicz W, Mirończuk AM. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl Microbiol Biotechnol. 2018;102:7669–78.
- [51]Eich A, Mildenberger T, Laforsch C, Weber M. Biofilm and diatom succession on polyethylene (PE) and biodegradable plastic bags in two marine habitats: Early signs of degradation in the pelagic and benthic zone? PLoS One. 2015;10(9):p.e0137201.