Effect of gentamicin and doxycycline on expression of relB and relE genes in Klebsiella pneumonia
Abstract
Klebsiella pneumoniae is responsible for a variety of disease in hospitalized patients. The goal of this study was to determine that K. pneumoniae isolates possessed toxin-antitoxin II genes such as relE and relB. Other than that, if there was a correlation between the expression of these two genes and antibiotic resistance in K. pneumoniae. Fifty-seven urine samples were collected from Baghdads’ hospitals; diagnosed and identified by phenotype and biochemical tests and confirmed with VITEK 2 compact system. Only fifteen isolates which were identified as Klebsiella pneumoniae. Antibiotic sensitivity was identified by using twelve antibiotics discs. K. pneumoniae showed 100% resistance to ceftriaxone, amoxicillin, ticarcillin, ticarcillin with clavulanic acid, ceftazidime, tetracycline, while other antibiotics showed less percent of resistant. Minimum inhibitory concentrations (MICs) of antibiotics detected by using macro tube dilution method to identify the antimicrobial activity for K. pneumoniae. The MIC of gentamicin and doxycycline antibiotics was 1024 Mg/ml, 512 Mg/ml, respectively. The relB (115 bp), and relE (136 pb) genes were detected by polymerase chain reaction. Then gene expression of relB and relE was conducted by using (RT-qPCR) technique treated with sub-MIC concentration of (gentamicin and doxycycline) antibiotics. This study found only ten isolates harbored the two genes. The relB gene expression was increased, but at the same time relE gene expression was decreased compared to control infB1 gene expression. This means the bacterial cell tolerance antibiotics sub-MIC concentrations by maintaining the number of bacteria under stress of antibiotics. Finally, these findings suggest the potential of relB to make K. pneumoniae resistant to antibiotics in their infections under antibiotic stress by the toxin-antitoxin II system.
INTRODUCTION
Klebsiella pneumoniae is a bacterial pathogen of major importance that causes a variety of disease manifestations in hospitalized patients [1]. K. pneumoniae is rapidly generating multidrug resistance (MDR), posing a severe hazard to patients due to a higher mortality rate and lower therapeutic efficiency. K. pneumoniae can develop antibiotic resistance more rapidly than other bacteria due to the production of enzymes such as extended-spectrum ß-lactamases (ESBLs) and carbapenemase [2-4]. Exposure to antibiotics is a major risk factor for developing antibiotic resistance in bacteria. The extensive and prolonged use of antibiotics is a crucial factor in the development of resistance in bacteria for diseases associated with healthcare. [5].
A toxin-antitoxin (TA) system is a group of two or more tightly related genes of a protein that encode a poison and a cure. In the conventional physiology of bacteria, an antitoxin attaches to a toxin and neutralizes it, preventing the bacterium from killing itself. This system consists of two genes in an operon, one of which produces a stable toxin and the other of which produces a less stable antitoxin [6].
The frequency of toxins-antitoxins system especially type II in bacteria, as well as their involvement in their pathogenicity, biofilm formation, and bacteriophage resistance in these bacteria which have toxin-antitoxin II system. The biological activities of these systems have many functions, including roles in antibiotic resistance and bacterial persistence [7].
This study aimed to determine whether or not K. pneumoniae isolates possessed toxins-antitoxins II genes such as relE and relB. Other than that, if there was a correlation between the expression of these two genes and antibiotic resistance in K. pneumoniae
MATERIALS AND METHODS
Bacterial isolation and identification
Fifteen K. pneumoniae were identified from fifty-seven samples of urine from patients by phenotyping and biochemical tests. The plates of MacConkey agar and blood agar were streaked with urine and then incubated at 37°C overnight. The bacteria showed pink colonies on MacConkey agar because of lactose fermentation. K. pneumoniae isolates showed the positive result for the Simmon citrate test indole test [8]. Then confirmation of identification was done by VITEK 2 Compact system.
Antibiotics sensitivity test
Resistant of isolates was determined using the disc’s technique [9] to twelve different antibiotics discs (Bioanalyse/Turkey): Ceftazidime(30µg), ceftriaxne (30µg), imipenem(10µg), amoxicillin (30µg), ciprofloxacin (10µg), ticarcillin (10µg), ticarcillin/clavulanic acid (75 µg /10µg), kanamycin(10µg), gentamicin (10µg) nitrofurantion (30µg), tetracycline(30µg), and doxycyciline (30µg). The isolated colony was cultured on nutrient broth overnight. Then it was cultured on Muller-Hinton agar after being diluted to 1.5×108 (cell/ml). Discs of antibiotics were fixed on the cultured plates by sterile forceps. After that, the plate was incubated at 37°C overnight. Then the results were compared with CLSI data in 2019 [10].
Antibiotic minimum inhibitory concentrations (MIC)
Isolates were tested for sensitivity by using the macro dilution broth assay, which was used to estimate the MIC of two antibiotics (doxycycline and gentamicin) for K. pneumoniae [9]. After dilution to 1.5×108 (cell/ml), bacteria were inoculated on Mueller-Hinton broth using a sterile loop, and antibiotics with double serial concentrations were administered to the medium. Then the medium was incubated for 24 hours at 37°C. The results of MIC value were the first clear test tube after turbid tubes.
PCR analysis
Thermal lysis and centrifugation method at 4°C for 30 seconds at 9,000 rpm were used to extract bacterial DNA. Nanodrop was used to assess the DNA content of the supernatant. Each step was completed in a final volume of 25 μl, by using 12.5 μl master mix, 1.5 μl primers and 100 ng DNA plus nuclease-free water. PCR amplification was used to identify relE and relB as previously described. The step of initial denaturation was done for 5 minutes at 95°C, while denaturation at 95°C for 30 sec. Then annealing was done at 62°C with the same time of previous step. After that the step of final extension was prepared at 72°C for 30 sec. Electrophoresis on 1.5% (w/v) agarose gel in tris acetate-EDTA (TBE) buffer resolved all PCR-amplified products. The samples were run at 100 V/Amp. for around 75 minutes. Using a conventional ultraviolet-light transilluminator, the specified products were detected following ethidium bromide staining [11]. The study genes were amplified with specific primers (Macrogen, USA) for relE: F-GCACTAAAGGAATGGCGAAAG, R-GGAGCTTGTTTGCTTCAATCC; relB: F-AATGGGCGTAACTCCTTCTG, R-CACAAGTTCAGCATCTTCATCAC; and infB1: F-CTCGCTGCTGGACTATATTCG, R-CGCTTTCAGCTCAAGAACTTC.
The same steps and conditions of PCR were used to investigate some toxin-antitoxin type II genes (relE and relB) and housekeeping genes. [12].
Gene expression of relE and relB genes by using Pfaffi method
The concentration of extracted RNA was measured using a Quantus Fluorometer to determine sample quality for downstream applications [13]. 199 µl of diluted quantus flour dye was combined with 1 µl of RNA. RNA concentrations were measured after a 5-minute incubation period at room temperature in a dark environment. Macrogen Company provided the primers in lyophilized form. As a stock solution, lyophilized primers were dissolved in nuclease-free water to a final concentration of 100 pmol/μl. 10 μl of primer stock solution was mixed with 90 μl of nuclease-free water.
RNA was isolated from K. pneumoniae using trizol reagent (Promega, USA) as described in the protocol by the manufacturer. Gene expression of toxin-antitoxin genes was measured via the relative (RT-qPCR) technique. The same conditions and program as performed previously by one-step real-time PCR with use the same primer sets of genes. The infB1 was used as housekeeping primer in real-time PCR with the same program steps but the annealing temperature was 50°C.
The gene expression was achieved with two positive isolates for these genes. And control isolate of K. pneumoniae without antibiotics treatment. Then analysis gene expression was calculated by using Pfaffi method for relative quantification as described previously [14, 15].
Folding change = 2-ΔΔCT
ΔCT = CT (gene) – CT (House Keeping gene)
ΔΔCT = ΔCT (Treated) – ΔCT (Control)
RESULTS
Effect of antibiotics on K. pneumoniae
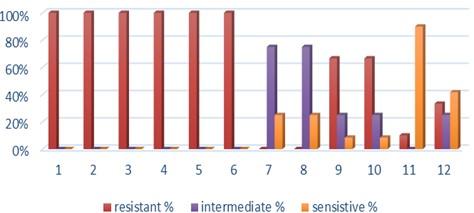
Fifteen K. pneumoniae were evaluated for antibiotic susceptibility to a variety of antibiotic types. K. pneumoniae isolates were 100 % resistant to ceftriaxone, amoxicillin, ticarcillin, ticarcillin with clavulanic acid, ceftazidime, and tetracycline, as shown in Figure 1. While ciprofloxacin and nitrofurantoin showed intermediate resistance (75%). While all isolates were sensitive to imipinem (94.6%) and only 6.4% were resitant to it. But doxycycline and gentamicin showed 67.7% of resistant isolates, and 32.3% of intermediate and persistent were sensitive. But in amikacin 35% of the isolates were sensitive and 25% were intermediate, and 40% were resistant.
Antibiotic minimum inhibitory concentrations
The macro tube dilution method was used to estimate the MIC for two antibiotics (doxycycline and gentamicin), when the first clear tube after serial of turbid tubes. The MICs value were 256 µg/ml for doxycycline and 1024 µg/ml for gentamicin as shows in Table 1.
Table 1. MICs value of antibiotics in K. pneumoniae.
Prevalence of relE and relB genes in the isolates
The prevalence of these genes in ten isolates is shown in Figure 2 and Figure 3. The results showed that only ten isolates were positive with products 136bp (relE) and 115bp (relB)) when 67 % of isolates have two genes relE and relB.
These results also confirmed the simultaneous presence of the toxin and antitoxin genes, as these genes are interrelated to neutralize the effect of the toxin on bacterial cells and prevent bacterial death.
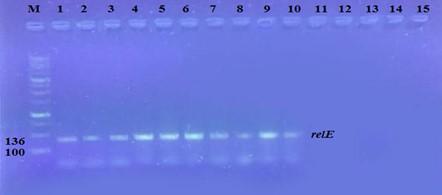
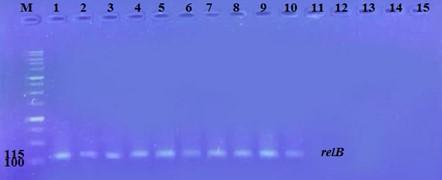
Effect of antibiotics on the expression of relE, and relB genes
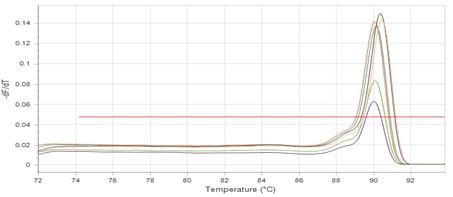
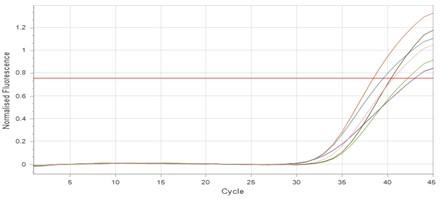
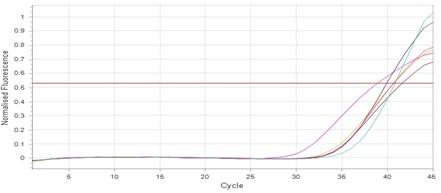
The results showed that RNA concentrations ranged from 31, 29, and 25 ng/ml in an untreated isolate (K2) to 26, 5, and 11 ng/ml in isolates treated with antibiotics with sub-MICs (doxycycline and gentamicin). The expression of the gene was detected by using RT- qPCR using specific primer (housekeeping gene of infB1). The amplification accuracy of gene products was noticed by the value of cycle threshold (Ct), as shown in Figure 3.
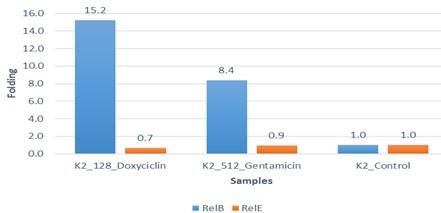
Due to the increasing incidence of novel resistant strains, it is critical to find inhibitors targeting K. pneumoniae to prevent infection by adopting alternate therapeutic strategies. The gene expression of relE and relB was studied after being treated with sub-MIC antibiotics. The data in Figures 4-6 showed that the relB gene was upregulated in treated isolates. The fold change in copy numbers between 8.4 and 15.2 was more than sixfold greater than the control. When reaching (0.7 to 0.9) copy numbers for relE gene in Figure 7.
DISCUSSION
K. pneumoniae is considered one of the most important bacteria because it causes many lethal diseases in the world. This study found that fifteen K. pneumoniae isolates were antibiotic resistant, with each isolate displaying a varied percentage of resistance. In another study from 801 samples, 580 were urine whereas, 221 were pus and sputum. Samples were collected from patients between 6 months to 90 years of both sexes [16]. Ten K. pneumoniae were isolated from urine and this outcome closely resembles the findings of the present study. They were 100% resistant to ampicillin; 70% to cefazoline, nitrofurantoin, and ofloxacin; 90% to cefotaxime and ceftriaxone; and 80% to ceftazidime. Imipenem followed by amikacin was the most effective antibiotic [17].
The genes relE and relB are type of toxin-antitoxin system type II and they are involved in antibiotics resistance [18]. Although 10 isolates harbored two genes from fifteen isolates, many studies mentioned that K. pneumoniae had these genes. Two hundred and twelve putative type II TA loci were identified in 30 replicons of these K. pneumoniae strains [19]. These results agree that the MIC concentration of the garlic extract can enhance the expression of the antitoxin gene since the bacteria preserve their numbers from death (programmed cell death) through decreasing gene expression of the toxin. This explains the possibility of using higher concentrations of MIC for used antibiotics [17]. The expression of the antitoxin relB might reverse the toxin relE. These findings showed that while the creation of the relE toxin does not kill cells, it does cause cell immobility when cells are exposed to antibiotics. The expression of the relB antitoxin might reverse this state of rest. Both genes are expressed under normal or favorable conditions, allowing the toxin’s effects to be inhibited. In contrast, the antitoxins are swiftly destroyed by proteases under stress, leaving the more stable toxin to impact cell development, generally as ribonucleases [18].
Another research looked at gene expression in the presence of gentamicin sub-MIC and relE1-relB1, hipA-hipB, doc-phd, and mazF-mazE loci were upregulated. Whereas the relE2-relB2 and vapC-vapB loci were downregulated. Since there is little information about the function of type II toxin-antitoxin systems in K. pneumoniae response to different stressors, the expression levels of TA system genes in K. pneumoniae were investigated under oxidative and antibiotic stress [20, 21]. These findings showed that bacterial cells can withstand antibiotic exposure due to a long-term upregulation of relE. This mechanism is unknown, but bacteria may use it to survive antibiotics and other stresses.
CONCLUSION
This study concluded that relB gene expression is downregulated in copy numbers compared to relE after being treated with antibiotics. This indicates the bacterial cell can tolerate antibiotics sub-MIC concentrations by maintaining their number under the stress of antibiotics. Thus, gentamicin and doxycycline antibiotics were used as a good treatment against K. pneumoniae. Further study is required on the toxin-antitoxin system and its function, as well as its relationship with antibiotic resistance in K. pneumoniae.
ACKNOWLEDGEMENT
The authors thank the technical support provided by Department of Biology / College of Science/University of Baghdad.
AUTHORS CONTRIBUTION
MTF; was designed the experiments; performed the experiments; analysis and recorded data: SEG and ZHS was conceived and designed the experiments, analyzed the data, guided to draft the manuscripts, and improved accordingly.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Fodah RA, Scott JB, Tam H-H, Yan P, Pfeffer TL, et al. (2014) Correlation of Klebsiella pneumoniae Comparative Genetic Analyses with Virulence Profiles in a Murine Respiratory Disease Model. PLoS ONE 9(9): e107394. doi: 10.1371/journal.pone.0107394.
- [2]Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998 Oct;11(4):589-603. doi: 10.1128/CMR.11.4.589.
- [3]Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019 Mar 1;43(2):123-144. doi: 10.1093/femsre/fuy043.
- [4]Munita JM, and Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2). https://doi.org/10.1128/microbiolspec.VMBF-0016-2015.
- [5]Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309-18. doi: 10.1179/2047773215Y.0000000030. Epub 2015 Sep 7. PMID: 26343252; PMCID: PMC4768623.
- [6]Van Melderen L, Bernard P, Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol. 1994 Mar;11(6):1151-7. doi: 10.1111/j.1365-2958.1994.tb00391.x.
- [7]Kamruzzaman M, Wu AY, Iredell JR. Biological Functions of Type II Toxin-Antitoxin Systems in Bacteria. Microorganisms. 2021 Jun 11;9(6):1276. doi: 10.3390/microorganisms9061276.
- [8]Collee JG, Miles RS, Watt B. Test for the identification of bacteria. In: Collee. J. G.; Fraser, A. G.; Marmion, B. P. and Simmons, A. (Eds.). Practical Medical Microbiology. 14P th P Edition. Churchill Livingstone, New York. pp. 131-146. 1996.
- [9]Kirby WMM, Bauer AW, Sherhis JC and Turck M. Antibiotic susceptibility testing by a standardized single disc method. Amer. J. Clin. Path. 1966; 45:493-496.
- [10]Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 29th ed. Clinical and Laboratory Standards Institute (CLSI). Supplement M100; 2019.
- [11]Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007 Feb;59(2):321-2. doi: 10.1093/jac/dkl481. Epub 2006 Dec 21. PMID: 17185300.
- [12]Narimisa N, Amraei F, Kalani BS, Mohammadzadeh R, Jazi FM. Effects of sub-inhibitory concentrations of antibiotics and oxidative stress on the expression of type II toxin-antitoxin system genes in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2020 June; 21:51-56. doi: 10.1016/j.jgar.2019.09.005. Epub 2019 Sep 11. PMID: 31520807.
- [13]chmittgen, T., Livak, K. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3 ;2008 :1101–1108. https://doi.org/10.1038/nprot.2008.73
- [14]Shehab ZH, AL-Rubaii BAL. Effect of D-Mannose on Gene Expression of Neuraminidase Produced from Different Clinical Isolates of Pseudomonas aeruginosa. Baghdad Science Journal.2019; 16(2):291-298. DOI: http://dx.doi.org/10.21123/bsj.2019.16.2.0291
- [15]Luma Saeed Mohammed, Enass Ghassan Sweedan, May Talib Flayyih. Effects of Alcoholic Extracts of Cinnamomum zeylanicum and Origanum Majorana on Expression of Hly Gene in Escherichia coli. Indian Journal of Forensic Medicine & Toxicology.2020; 14(3):937-942.
- [16]Sweedan Enass Ghassan. Estimate Antimicrobial activity and Antibiofilm formation of bark (Cinnamomum zeylanicum) on Klebsiella pneumoniae from urinary tract infections. Iraqi Journal of Science. 2018; 59(3C): 1560-1566. DOI:10.24996/ijs.2018.59.3C.3
- [17]Fernández-García L, Blasco L, Lopez M, Bou G, García-Contreras R, Wood T, Tomas M. Toxin-Antitoxin Systems in Clinical Pathogens. Toxins (Basel). 2016 Jul 20;8(7):227. doi: 10.3390/toxins8070227. PMID: 27447671; PMCID: PMC4963858.
- [18]Zina Hashem Shehab, Enass Ghassan Sweedan, May Talib Flayyih. Evaluation the effect of Allium sativum (garlic) oil on the expression of mazE and mazF genes in Escherichia coli clinical isolates. Biochem. Cell. Arch. 2021;21(1):721-726.
- [19]Wei YQ, Bi DX, Wei DQ, Ou HY. Prediction of Type II Toxin-Antitoxin Loci in Klebsiella pneumoniae Genome Sequences. Interdiscip Sci. 2016 Jun;8(2):143-149. doi: 10.1007/s12539-015-0135-6. Epub 2015 Dec 10. PMID: 26662948.
- [20]Lieven Buts, Jurij Lah, Minh-Hoa Dao-Thi, Lode Wyns, and Remy Loris. Toxin–antitoxin modules as bacterial metabolic stress managers. TRENDS in Biochemical Sciences.2005;30 (12):672-679.
- [21]Negar Narimisa,Fatemeh Amraei, Behrooz Sadeghi Kalani. Biofilm establishment, biofilm persister cell formation, and relative gene expression analysis of type II toxin-antitoxin system in Klebsiella pneumoniae. Gene Reports. 2020; 21(2):100846. DOI: 10.1016/j.genrep.2020.100846.