MicroRNA-221-3p promotes cell proliferation, migration, and invasion in gastric cancer by modulating PIK3R1
Abstract
Gastric cancer (GC), which is the fourth most prevalent cancer in the world is significantly threatened the health of people, particularly those in developing nations. Nearly all significant pathological and physiological mechanisms, including apoptosis, proliferation, cell cycle, differentiation, as well as DNA damage, are regulated by miRNAs. This study investigated the miR-221-3p expression and identified its target genes in GC tissue samples and cell lines, for an understanding of the miR-221-3p influence and basic processes in the progression of GC. GC tissues and matched marginal tissues were taken from 50 patients undergo gastric surgery. MiR-221-3p mimics, inhibitors, and negative controls (NC) were transfected into MKN-45 cells, using Lipofectamine RNAiMAX reagent. The proliferation was assessed by the MTT assay. Cell migration and invasion was assessed by Transwell assay. By combining Western blotting and qRT-PCR, the impact of miR-221-3p in the PIK3R1 expression in gastric cancer cells was examined. Overexpression of miR-221-3p significantly enhanced the migration, invasion, and proliferation of gastric cancer cells, conversely, transfection of miR-221-3p inhibitor led to opposite effect caused by overexpression of this miRNA on phenotypic characteristics of gastric cancer cell line. Additional investigation revealed that PIK3R1 was downregulated significantly by overexpression of miR-221-3p. Whereas, when the MKN-45 cells transfected with miR-221-3p inhibitor, PIK3R1 was noticeably overexpressed. Our current data indicate that miR-221-3p possibly work as a tumor promoter in the development of gastric cancer by negatively regulating PIK3R1 expression, hence miR-221-3p/ PIK3R1 highlighted as promising therapeutic targets or prognostic and diagnostic biomarkers for GC patients.
INTRODUCTION
Gastric cancer (GC), which is the fourth most prevalent cancer in the world is significantly threatened the health of people, particularly those in developing nations [1,2]. The prognosis of GC is significantly influenced by early detection and effective therapy [3–5]. Several genes, factors, and signals are involved in the formation of GC [6,7]. At present, the standard methods for diagnosing cancer are histopathological, morphological, and immunological tests [8,9]. Genetic diagnostics particularly emphasizes on cellular alterations and seeks to spot early signs of cancer in normal cells, has emerged as a result of advancements in medical technology [5].
MicroRNA are species of noncoding endogenous RNA-type molecules (about 19-25 nucleotides) that are produced by RNA polymerases II (Pol II) and post-transcriptionally regulates the expression of target mRNAs via degradation or translational repression [10,11]. The main role of miRNA is negatively regulating gene expression by silencing (blocking) the messenger RNA or degrading it via binding direct to 3′ (UTR) regions of the target mRNAs [12–14]. Nearly all significant pathological and physiological mechanisms, including drug resistance, apoptosis, proliferation, cell cycle, metabolism, differentiation, as well as DNA damage, are regulated by miRNAs [15–17]. As a result, any abnormal expression or deregulation of miRNA function may result in the emergence of pathogenic events, including gastric cancers.
Recent investigations have revealed that the miR-221-3p, which is a mature type of the miR-221, is markedly up-regulated in cervical cancer [18], breast cancer [19], colorectal cancer [20] and GC [21]. MiR-221-3p has also been demonstrated to control aberrant cellular differentiation and proliferation in a number of malignancies [18,20,22,23], despite the fact that few research have investigated it in relation to GC [21,24]. MiR-221-3p overexpression is frequently seen in the majority of tumors, however the exact mechanism underlying this overexpression is yet unknown. Therefore, there is a definite need for more investigations into the particular genes that this miRNA targets in GC as well as for the development of efficient therapies that target this important genetic regulator.
The PIK3R1 gene, which codes for p85, p55, and p50, is found on the 5q13.1 region of the human genome. According to the TCGA, PIK3R1 is the 11th most frequently altered gene among all cancer’s lineages [25]. When compared to healthy control tissues, PIK3R1 is dramatically downregulated in a range of human cancerous tissues, including lungs, prostate, hepatic, kidneys, breast, and cervical cancer [26–28]. Additionally, it has been demonstrated that numerous miRNAs implicated in the control of cancer growth have PIK3R1 as a functional target [29]. According to these results, PIK3R1 expression deregulation is a major factor in the emergence of cancer. The purpose of the present study looked at the miR-221-3p expression and to identify its target genes in GC tissue specimens and cell lines, to comprehend the miR-221-3p influence in the development of GC.
MATERIALS AND METHODS
Patients and gastric cancer tissue samples
From the surgical specimens, 100 pairs of GC tissues and matched marginal tissues were taken from 50 patients having gastric surgery. After being histologically verified, tumor tissues were stored in a -80°C freezer with their matched marginal tissues. Patients received neither radiotherapy nor chemotherapy before surgery. The study was approved by the research committee of the medical ethics unit, university of Tabriz (978661142). A written consent obtained from the eligible participants to be admitted in the study and based on their desire to participate in the research project.
Cell line
The MKN-45 gastric cancer cells were purchased from Procell life Science and technology (Wuhan, China). RPMI 1640 medium (HiMedia Laboratories, India) supplemented with 10% FBS, 100 U/ml penicillin and 0.1 mg/ml streptomycin was used to cultivate cells and incubated at 37°C in a 5% CO2 humid environment.
Cells transfection
MiR-221-3p mimics or NC and miR-221-3p inhibitors or NC were transfected into MKN-45 cells, using Lipofectamine RNAiMAX reagent (Invitrogen, USA). 6-well plates were used for MKN-45 cell culture, following 60–80% confluent, the manufacturer's directions were followed for transfection. miR-221-3p mimics, inhibitor, and negative control (NC) were purchased from BIONEER Corporation (BIONEER Corporation, Korea). The final concentrations were 10 nmol.
Total RNA extraction
Total RNA extraction from frozen tissues samples and transfected cell line 24h after transfection was done by FavorPrep tissue total RNA kit (Favorgen Biotech Inc., Taiwan) and according to manufacturer's instructions. Nano-Drop 2000 Spectrophotometer (Thermo-Fischer Scientific, USA) was used to determine the concentration of RNAs.
Quantitative real time PCR
First strand complementary DNA from the total RNA was prepared by using of HyperScript™ cDNA synthesis Kit (GeneAll Biotechnology, Korea), and as manufacturer instructions. Quantitative PCR was used to examine miR-221-3p and PIK3R1 expression by utilizing SYBR Green master mix. (Geneall, Korea). The miRprimer2 program and sRNAPrimerDB web tool were used to create specific primer for miR-221-3p by utilizing miRBase database to get sequence of microRNA, while NCBI primer designing tools was used to design (PIK3R1) primer and all the primers were synthesized by Macrogen (Macrogen, Co., Korea), U6 and GAPDH were employed as controls. The primer sequences utilized in the present study are given in (Table 1). The data results of qRT-PCR were analyzed using relative quantification method by using of 2-ΔΔCt [30].
Table 1. Primers details used in the current study.
Bioinformatics analysis for target gene prediction
Bioinformatics databases (TargetScan, miRecords and miRDB) were used to detect the miR-221-3p binding sequence at the 3'-UTR of PIK3R1. We obtained predictive gene from these websites databases to improve the reliability of the prediction.
Western blot analysis
The primary antibody against PIK3R1 was selected for this study (Fine Biotech co., Ltd, China), the secondary antibodies were obtained from (Elabscience, China) and GAPDH (Fine Biotech co., Ltd, China) was employed as the control. Extraction of total protein from cells was done by Total Protein Extraction Kit (Elabscience, China) 72 h after transfection. 12% SDS-PAGE was used to separate protein from each sample and then transferred to (PVDF) membranes in an equal proportion. Membranes were soaked with TBST Buffer (containing 5% Skim Milk) as blocking buffer and the membrane blocked at room temperature for 1.5h then membranes with primary antibodies incubated overnight at 4°C with gentle shaking. Following incubation, membranes were then incubated at room temperature with HRP-conjugated secondary antibodies for 1 hour on a shaker. By using ECL reagents, the bands were identified and subjected to X-rays for imaging.
Cell proliferation assay
Following treatment of cells with mimics or inhibitors, 96-well plates was used to seed MKN-45 cells at a density of 2000 cells/well, then cells were incubated in 5% CO2 at 37 ˚C for periods of 24 hours and 48 hours, respectively. To each well a 10 μl of MTT solution (Fine Biotech co., Ltd, China) was added. Cells were incubated for an additional period of 3-4 hours at 37 ˚C, and 570 nm was used to measure the absorbance.
Cell migration and invasion assays
Cell migration was assessed by utilizing the Transwell technology (Sigma-Aldrich, UK). Briefly, 2x105 transfected MKN-45 cells in media without serum were placed on the top compartment of Transwell chamber without Matrigel. As a chemoattractant, 10% FBS-supplemented DMEM medium was introduced to the lower compartment. After that, the plate incubated for 16-48h at 37 °C. Following the incubation time, a cotton swab was used to wipe cells in the top chamber. Then, 0.2% crystal violet was used to fix, and stain migrated or invaded cells to the lower layer of the Transwell chamber and four random fields for each well were chosen to count cells. Cell invasion was assessed with Transwell system (Sigma-Aldrich, UK) in the same procedure of migration with only difference that transfected cells were added to the top compartment coated with Matrigel.
Statistical analysis
All statistical analysis was done by SPSS version 21 (IBM Corporation, USA). Mean ± SD was used to display results. Paired t test and one-way ANOVA were utilized to calculate differences between two or more groups. A statistically significant were set as *p¬< 0.05 and **¬p< 0.01, respectively.
RESULTS
miR-221-3p expression in gastric cancer tissue samples and cell lines
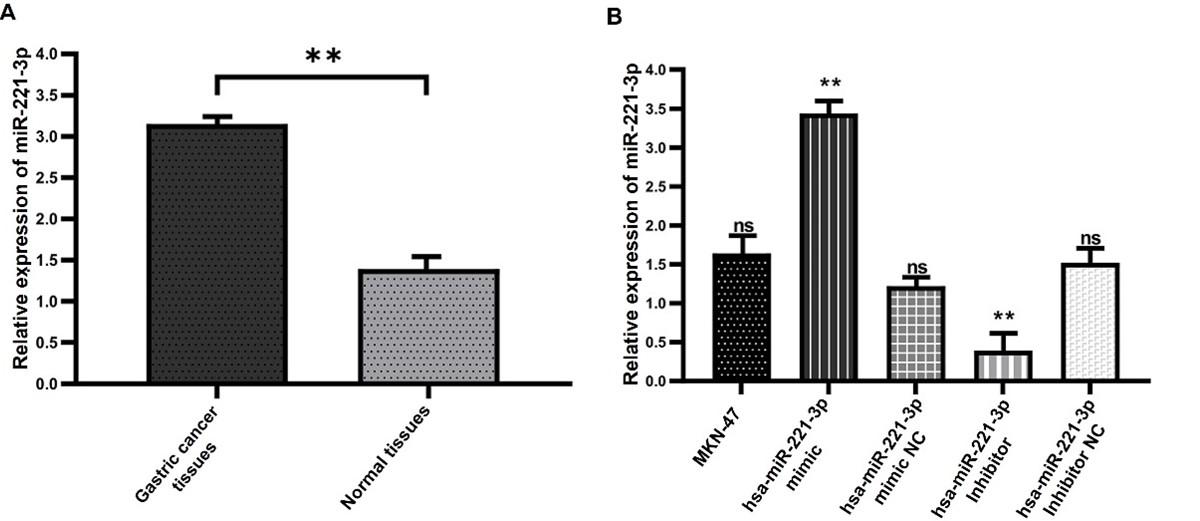
QRT-PCR has been used to identify the miR-221-3p expression level in cell lines and tissue samples. The findings demonstrated that gastric cancer tissues exhibited a significant expression level of miR-221-3p greater than adjacent non-tumorous tissues (Figure 1A). Then, RNA was extracted before transfection and 24 hours post miR-221-3p mimics and inhibitor transfected to MKN-45 cells. In comparison to control (cell line with no transfection), the expression of miR-221-3p significantly elevated after cells were transfected with miR-221-3p mimic, whilst in miR-221-3p inhibitor group the expression of miR-221-3p significantly reduced. No alterations in the expression were noticed between control and miR-221-3p mimics NC or miR-221-3p inhibitor NC groups (Figure 1B).
miR-221-3p enhanced the proliferation, migration and invasion of MKN-45 cells
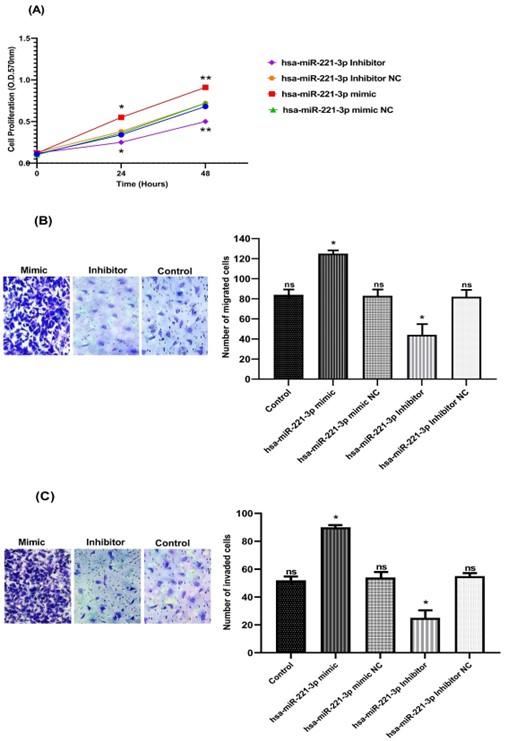
To additional assess miR-221-3p effects on proliferation in gastric cancer cell line, MTT assay was used. The data revealed that the proliferation of MKN-45 cells significantly reduced after miR-221-3p knocking down in comparison to control. On the other hand, miR-221-3p overexpression had the opposite outcome and exhibited a significant increasing of proliferation in MKN-45 cells in comparison to control, recent findings indicating that miR-221-3p significantly enhances MKN-45 cells proliferation (Figure 2A). Transwell invasion and migration assay demonstrated that cells transfected with miR-221-3p inhibitor exhibited a significant decrease in migratory and invasive cells number in comparison to control, while miR-221-3p mimics group displayed a significant increase in migratory and invasive cells number in comparison to control. No significant differences were observed in cellular migration and invasion between all control groups (Figure 2B and C).
miR-221-3p modulated endogenous PIK3R1 expression of MKN-45 cells
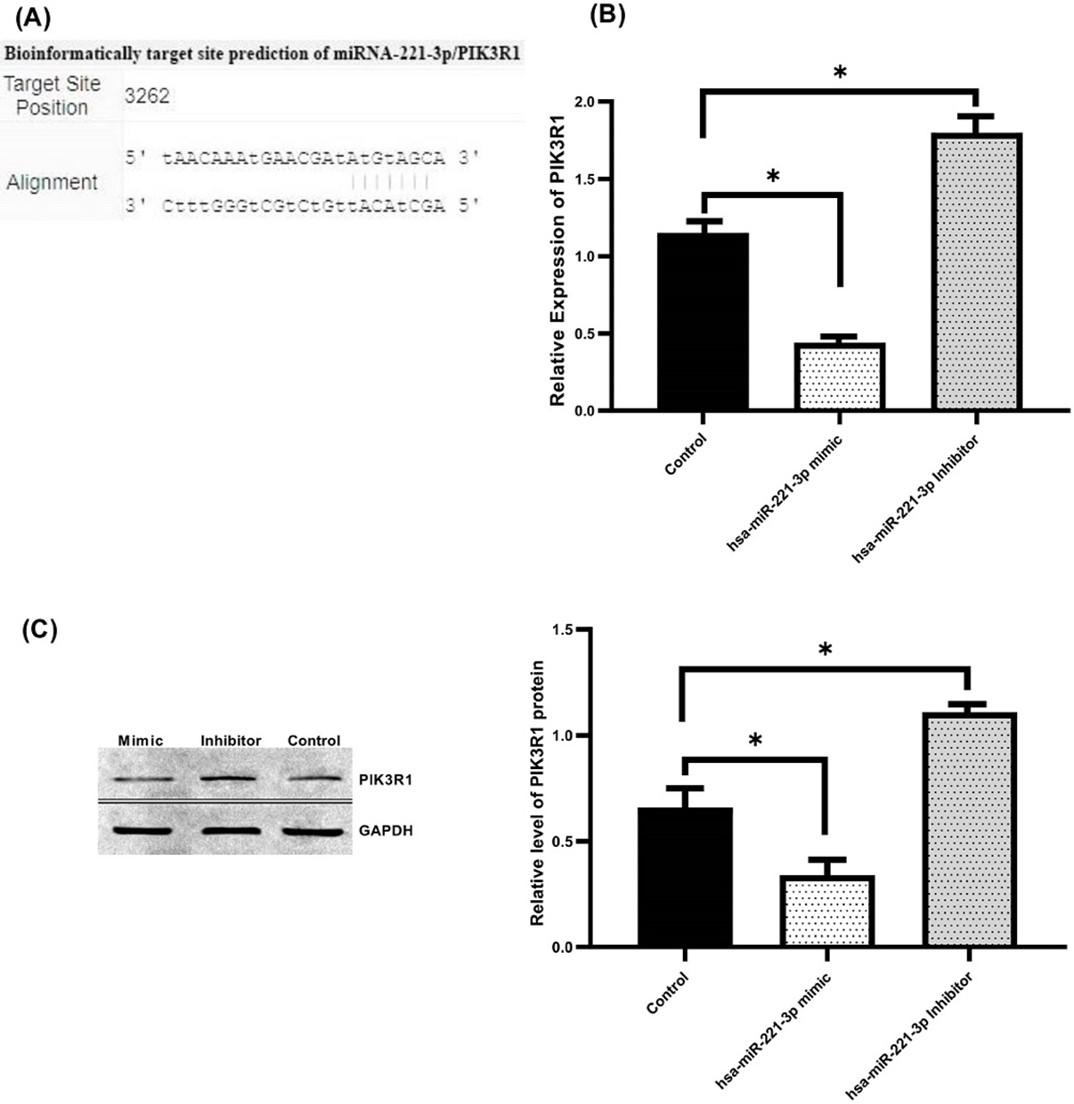
Since the experiments on cell function were highlighted and indicated that miR-221-3p enhance the proliferation, migration, and invasion of GC cell. We assumed that miR-221-3p might affect the development and occurrence of GC via a particular molecular mechanism. Thus, bioinformatics analysis was used to study how the cell proliferation, migration, and invasion are regulated by miR-221-3p through using (TargetScan, miRecords and miRDB) online tool to found possible interaction of PIK3R1 with miR-221-3p. Results of our study showed that miR-221-3p had the potential binding sequence targeting 3’-UTR of PIK3R1 (Figure 3A), also PIK3R1 and its relation to gastric cancer consider one of the less studied genes. Therefore, we chose PIK3R1 to further examine using qRT-PCR and Western blotting. Our findings revealed that PIK3R1 expression was significantly dampened in mimic group at the mRNA (Figure 3B) and protein level (Figure 3C) in comparison to control group. Conversely, a significant increase in the PIK3R1 expression was noticed in MKN-45 cells following knock-down of miR-221-3p at the mRNA and protein level relative to the control group. Overall, our findings imply that miR-221-3p negatively modulates PIK3R1.
DISCUSSION
Numerous genes expression profiling studies have discovered miRNAs linked to GC. Including miR-106a-5p and miR-21-5p that were discovered to be overexpressed in the tissues of GC [31–34], also miR-497-5p, miR-16-5p, miR-335-3p and let-7c-5p, that downregulated in tissues from GC [35–39]. In agreement with previous studies [40,41], our data revealed that gastric cancer tissue samples had miR-221-3p levels greater than matched non-tumorous. In relation to the above studies, gastric cancer has been verified to have higher contents of miR-221-3p, pointing a possible function for miR-221-3p as oncogenic in GC which is compatible with our findings.
In the present study, a number of cell investigations were established to show the molecular influence of miR-221-3p on GC. Investigations of miR-221-3p transfected cells demonstrated that overexpression of miR-221-3p enhanced proliferative effect in MKN-45 cells, in comparison to control. Moreover, a significant enhancing of migratory and invading abilities was shown in MKN-45 cell line transfected with miR-221-3p mimics, suggesting a strong promoting role of miR-221-3p in cells’ migration and invasive ability. Conversely, miR-221-3p knockdown in MKN-45 cells led to the opposite effect caused by overexpression of this miRNA on phenotypic characteristics of gastric cancer cell line.
A similar observation on how miR-221-3p affects tumorigenesis has been studied in many cancers, for example, miR-221-3p overexpression enhance proliferation of pancreatic cancer cell via PTEN-Akt pathway [42]. Also, it was established that the invasion, migration, and proliferation of AML2 cells were induced by miR-221-3p and promoted the cell cycle arrest in G1/S phase also cell apoptosis inhibition through targeting CDKN1C and revealed that miR-221-3p stimulates acute myelocytic leukemia progression [43]. Through targeting LIFR, overexpression of miR-221-3p facilitated HCC cells migration, invasion, and proliferation [44]. In addition, miR-221-3p was strongly expressed and enhanced cell invasion and proliferation by targets and downregulates PTEN in gastric cancer cells [45].
To clarify how miR-221-3p conflicts functions in various tumor types, we discovered that miR-221-3p targets several genes of which PIK3R1 gene and we found that miR-221-3p can alter PIK3R1expression. Thus, we conducted additional studies using Western blotting and qRT-PCR and discovered that PIK3R1 expression was downregulated significantly at the mRNA and also protein levels in miR-221-3p transfected MKN-45 cells compared to control group. Conversely, a significant increase in the PIK3R1 expression at the mRNA and also protein levels was noticed in MKN-45 cells following knock-down of miR-221-3p in comparison to control group.
The regulatory subunit of type I PI3K (p85α) is encoded by PIK3R1. Numerous biological processes are under the control of the PI3K/AKT pathway, including migration, invasion, and proliferation [46,47]. The PI3K pathway is downstream activated when the p85α protein is lost [48]. Many different cancers, including pancreatic cancer, have been reported to express PIK3R1 aberrantly [49]. PIK3R1 expression contributes to kidney cancer cell migration and proliferation [50]. Additionally, breast cancer cells are more likely to proliferate, migrate, and invade when miR-21 directly targets PIK3R1 and activates the PI3K/AKT signaling pathway [51]. Another study revealed that PIK3R1 was extremely expressed in HCC tissues which enhanced migration, proliferation as well as inhibited apoptosis of HCC cell lines [52].
Furthermore, it is recognized that PI3K/Akt/mTOR pathway plays a part in a number of biological and important cellular processes, including metastasis, apoptosis, and proliferation [53,54]. Evidence suggests that PI3K/Akt/mTOR pathway inhibitors may treat GC by inducing apoptosis, reducing cell proliferation, and improving chemotherapy sensitivity [54]. AKT is frequently overactivated in malignant tumors due to PIK3R1 inactivation, which also increases invasion and migration while decreasing apoptotic susceptibility.
Therefore, the level of PIK3R1 in both protein and mRNA suggested that it played significant roles in the development of GC. These findings, when combined with the previous data, strongly supported the PIK3R1 functions as tumor suppressor in GC. Generally, PIK3R1 is an attractive miR-221-3p target gene in GC. On the one hand, PIK3R1 is a crucial part of the PI3K/Akt pathway that is recognized as a major factor in the development of GC. The present study revealed that miR-221-3p can bind to PIK3R1 in GC cells, also miR-221-3p expression can influence PIK3R1expression. Thus, gastric cancer progression and occurrence may be impacted by miR221-3p through influencing and regulating the expression of PIK3R1 gene and the PI3K/AKT signal pathway which potential mechanism underlies the oncogenic properties of miR-221-3p and indicating that PIK3R1 might be a therapeutic target and a prognostic factor for GC.
In summary, studies on how miR-221-3p regulates PIK3R1 and its participation in the growth and metastasis of gastric cancer cells will help us better understand how miR-221-3p promotes tumorigenesis and provide insight into the mechanisms and progression of gastric cancer. These findings will give a clear theoretical framework for the detection and management of GC. Our exploratory research presented that mir-221-3p and PIK3R1 have regulatory interactions at the vitro levels; though, in vivo investigation needs additional examined by specific experimental consequences.
CONCLUSIONS
Our study has limitation due to the small sample size, as well as the high cost and difficulty in obtaining some materials and kits also lack of studies related to this subject, so more experimental studies are required. Despite this, our investigations pointed an interesting correlation between miR-221-3p and PIK3R1 in GC occurrence and development as miR-221-3p promoting GC cell proliferation, invasion and migration indicating that miR-221-3p serves as tumor promoter via negatively regulating PIK3R1. Thus, these findings point to a promising therapeutic and diagnostic role of miR-221-3p/PIK3R1 in GC, which may help us understand the mechanisms behind the etiology of GC. In addition, future studies are required to validate the miR-221-3p binding sequence targeting the 3'-UTR and how regulates PIK3R1.
ACKNOWLEDGEMENT
We would like to thank the patients for their cooperation in giving samples to complete this project; also, we are thankful to the staff of the laboratory, for technical support. The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
AUTHOR CONTRIBUTIONS
All authors had equal roles in design, work, statistical analysis, and manuscript writing.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Alessandrini L, Manchi M, De Re V, Dolcetti R, Canzonieri V. Proposed Molecular and miRNA Classification of Gastric Cancer. Int J Mol Sci 2018;19:1683.
- [2]Backes C, Meese E, Keller A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol Diagn Ther 2016;20:509–18.
- [3]Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700– 13.
- [4]Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597–610.
- [5]Wang C, Huang Y, Zhang J, Fang Y. MiRNA-339-5p suppresses the malignant development of gastric cancer via targeting ALKBH1. Exp Mol Pathol 2020;115:104449.
- [6]Molaei F, Forghanifard MM, Fahim Y, Abbaszadegan MR. Molecular Signaling in Tumorigenesis of Gastric Cancer. Iran Biomed J 2018;22:217–30.
- [7]Pasechnikov V. Gastric cancer: Prevention, screening and early diagnosis. World J Gastroenterol 2014;20:13842.
- [8]Salati M, Orsi G, Smyth E, Aprile G, Beretta G, De Vita F, et al. Gastric cancer: Translating novels concepts into clinical practice. Cancer Treat Rev 2019;79:101889.
- [9]Shin VY, Chu K-M. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol 2014;20:10432–9.
- [10]Yu X, Wang M, Li L, Zhang L, Chan MTV, Wu WKK. MicroRNAs in atopic dermatitis: A systematic review. J Cell Mol Med 2020;24:5966–72.
- [11]Sohel M. Choice of samples in extracellular microRNA research: Which fraction is better- exosomal or nonexosomal? J Adv Biotechnol Exp Ther 2018;1:11.
- [12]Zhou C, Zhao X, Duan S. The role of miR‐543 in human cancerous and noncancerous diseases. J Cell Physiol 2021;236:15–26.
- [13]Liu X, Ma R, Yi B, Riker AI, Xi Y. MicroRNAs are involved in the development and progression of gastric cancer. Acta Pharmacol Sin 2021;42:1018–26.
- [14]Mawlah Y, Naji M, Imari M, Abdulabbas H. Micro-RNA evaluation, specification, and stabilization study in mixed/non-mixed body fluids as a specific molecular marker. J Adv Biotechnol Exp Ther 2022;5:347.
- [15]Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22–36.
- [16]Abedi F, Rezaee R, Hayes AW, Nasiripour S, Karimi G. MicroRNAs and SARS-CoV-2 life cycle, pathogenesis, and mutations: biomarkers or therapeutic agents? Cell Cycle 2021;20:143–53.
- [17]Liu F, Bu Z, Zhao F, Xiao D. Increased T-helper 17 cell differentiation mediated by exosome-mediated microRNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci 2018;109:65–73.
- [18]Wu Q, Ren X, Zhang Y, Fu X, Li Y, Peng Y, et al. MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem Biophys Res Commun 2018;497:1162–70.
- [19]Deng L, Lei Q, Wang Y, Wang Z, Xie G, Zhong X, et al. Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget 2017;8:108712–25.
- [20]Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, et al. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res 2014;6:391–401. PMID: 25075256; PMCID: PMC4113501.
- [21]Shi J, Zhang Y, Jin N, Li Y, Wu S, Xu L. MicroRNA-221-3p Plays an Oncogenic Role in Gastric Carcinoma by Inhibiting PTEN Expression. Oncol Res Feat Preclin Clin Cancer Therap 2017;25:523–36.
- [22]Xue Y, Wei Z, Ding H, Wang Q, Zhou Z, Zheng S, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1α in the progression of atherosclerosis. Atherosclerosis 2015;241:671–81.
- [23]Ergun S, Tayeb TS, Arslan A, Temiz E, Arman K, Safdar M, et al. The investigation of miR-221-3p and PAK1 gene expressions in breast cancer cell lines. Gene 2015;555:377–81.
- [24]Zhang Y, Huang H, Zhang Y, Liao N. Combined Detection of Serum MiR-221-3p and MiR-122-5p Expression in Diagnosis and Prognosis of Gastric Cancer. J Gastric Cancer 2019;19:315.
- [25]Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4.
- [26]Lin Y, Yang Z, Xu A, Dong P, Huang Y, Liu H, et al. PIK3R1 negatively regulates the epithelialmesenchymal transition and stem-like phenotype of renal cancer cells through the AKT/GSK3β/CTNNB1 signaling pathway. Sci Rep 2015;5:8997.
- [27]Munkley J, Livermore KE, McClurg UL, Kalna G, Knight B, McCullagh P, et al. The PI3K regulatory subunit gene PIK3R1 is under direct control of androgens and repressed in prostate cancer cells. Oncoscience 2015;2:755–64.
- [28]Wang Y, Chen A, Zheng C, Zhao L. miR‐92a promotes cervical cancer cell proliferation, invasion, and migration by directly targeting PIK3R1. J Clin Lab Anal 2021;35.
- [29]Pan X, Hong X, Lai J, Cheng L, Cheng Y, Yao M, et al. Exosomal MicroRNA-221-3p Confers Adriamycin Resistance in Breast Cancer Cells by Targeting PIK3R1. Front Oncol 2020;10.
- [30]Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001;25:402–8.
- [31]Xiao B, Guo J, Miao Y, Jiang Z, Huan R, Zhang Y, et al. Detection of miR-106a in gastric carcinoma and its clinical significance. Clinica Chimica Acta 2009;400:97–102.
- [32]Sun Y, Chen G, He J, Huang Z-G, Li S-H, Yang Y-P, et al. Clinical significance and potential molecular mechanism of miRNA-222-3p in metastatic prostate cancer. Bioengineered 2021;12:325–40.
- [33]Fu P, Lin L, Zhou H, Zhao S, Jie Z. Circular RNA circEGFR regulates tumor progression via the miR106a-5p/DDX5 axis in colorectal cancer. Braz J Med Biol Res 2021;54.
- [34]Dong S, Zhang X, Liu D. Overexpression of long noncoding RNA GAS5 suppresses tumorigenesis and development of gastric cancer by sponging miR-106a-5p through the Akt/mTOR pathway. Biol Open 2019.
- [35]Feng L, Cheng K, Zang R, Wang Q, Wang J. miR-497-5p inhibits gastric cancer cell proliferation and growth through targeting PDK3. Biosci Rep 2019;39.
- [36]Zhang L, Yao L, Zhou W, Tian J, Ruan B, Lu Z, et al. miR-497 defect contributes to gastric cancer tumorigenesis and progression via regulating CDC42/ITGB1/FAK/PXN/AKT signaling. Mol Ther Nucleic Acids 2021;25:567–77.
- [37]Jiang X, Wang Z. miR-16 targets SALL4 to repress the proliferation and migration of gastric cancer. Oncol Lett 2018.
- [38]Zhang L, Wu J, Ling MT, Zhao L, Zhao K-N. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol Cancer 2015;14:87.
- [39]Zhu C, Huang Q, Zhu H. Melatonin Inhibits the Proliferation of Gastric Cancer Cells Through Regulating the miR-16-5p-Smad3 Pathway. DNA Cell Biol 2018;37:244–52.
- [40]Chun-zhi Z, Lei H, An-ling Z, Yan-chao F, Xiao Y, Guang-xiu W, et al. MicroRNA-221 and microRNA222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010;10:367.
- [41]Cai H, Yuan Y, Hao Y-F, Guo T-K, Wei X, Zhang Y-M. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol 2013;30:452.
- [42]Yang W, Yang Y, Xia L, Yang Y, Wang F, Song M, et al. MiR-221 Promotes Capan-2 Pancreatic Ductal Adenocarcinoma Cells Proliferation by Targeting PTEN-Akt. Cell Physiol Biochem 2016;38:2366–74.
- [43]Zhang X, Xu Y, Wang J, Zhao S, Li J, Huang X, et al. miR-221-3p Delivered by BMMSC-Derived Microvesicles Promotes the Development of Acute Myelocytic Leukemia. Front Bioeng Biotechnol 2020;8
- [44]Tan W, Li Z, Xia W, Zhu J, Fan R. miR-221-3p regulates hepatocellular carcinoma cell proliferation, migration and invasion via targeting LIFR. Ann Hepatol 2022;27:100567.
- [45]Shi J, Zhang Y, Jin N, Li Y, Wu S, Xu L. MicroRNA-221-3p Plays an Oncogenic Role in Gastric Carcinoma by Inhibiting PTEN Expression. Oncol Res Feat Preclin Clin Cancer Therap 2017;25:523–36
- [46]Chen L, Yang L, Yao L, Kuang X-Y, Zuo W-J, Li S, et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun 2018;9:1357.
- [47]Kim S, Lee E, Jung J, Lee JW, Kim HJ, Kim J, et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene 2018;37:2982–91.
- [48]Toste PA, Li L, Kadera BE, Nguyen AH, Tran LM, Wu N, et al. p85α is a microRNA target and affects chemosensitivity in pancreatic cancer. J Surg Res 2015;196:285–93.
- [49]Kong Y, Li Y, Luo Y, Zhu J, Zheng H, Gao B, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer 2020;19:82.
- [50]Wang Y-D, Sun Z-L. Effects of miR-455 on PIK3R1 gene expression regulation and kidney cancer cell functions. Eur Rev Med Pharmacol Sci 2017;21:3370–6.
- [51]Yan L, Liu Y, Xiang J, Wu Q, Xu L, Luo X, et al. PIK3R1 targeting by miR-21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT, and predicts clinical outcome of breast cancer. Int J Oncol 2016;48:471–84..
- [52]Ai X, Xiang L, Huang Z, Zhou S, Zhang S, Zhang T, et al. Overexpression of PIK3R1 promotes hepatocellular carcinoma progression. Biol Res 2018;51:52.
- [53]Willems L, Tamburini J, Chapuis N, Lacombe C, Mayeux P, Bouscary D. PI3K and mTOR Signaling Pathways in Cancer: New Data on Targeted Therapies. Curr Oncol Rep 2012;14:129–38.
- [54]Matsuoka T, Yashiro M. The Role of PI3K/Akt/mTOR Signaling in Gastric Carcinoma. Cancers (Basel) 2014;6:1441–63.