Genome-wide identification and characterization of interleukin-18 gene family in rainbow trout (Oncorhynchus mykiss)
Abstract
Rainbow trout (Oncorhynchus mykiss) are cold freshwater fish species belonging to the Salmonidae family, and they hold significant economic importance in the global aquaculture industry. The interleukin-18 (IL-18) gene family plays a crucial role in the immune response of rainbow trout when combating viral infections. A wide range of computational approaches were conducted to investigate the functions, phylogeny, and expressions of IL-18 family genes. The study found that there were nine IL-18 genes present on eight chromosomes of rainbow trout, and their encoded proteins were predicted to be distributed in the cytoplasm and nucleus. Genetic structures, motif and conserved domains analysis revealed that IL-18 genes in the same group have similar exons, motifs, and conserved domains. The phylogenetic study demonstrates the divergence of 38 IL-18 genes from three fish species into three distinct subgroups across time. The synteny analysis revealed the evolutionary mechanisms, such as gene duplications and mutations. In addition, the research investigated the expression of IL-18 genes in the liver tissues of rainbow trout following infection with infectious hematopoietic necrosis virus. These findings provide a comprehensive understanding of the role of IL-18 genes in O. mykiss in response to pathogenic infection.
INTRODUCTION
Rainbow trout (Oncorhynchus mykiss) holds substantial economic value and fulfills a critical role in the global seafood supply chain [1]. It stands out as a highly adaptable and suitable species for production; however, the rapid increase in rainbow trout production has also given rise to several diseases [2, 3]. Among them, viral diseases pose a significant threat to major aquaculture salmonid species O. mykiss [4]. For example, infectious pancreatic necrosis (IPN), pancreatic disease (PD), infectious hematopoietic necrosis (IHN), viral hemorrhagic septicemia (VHS), and infectious salmon anemia (ISA) cause substantial economic setbacks [4, 5]. Interestingly, there are only a limited number of effective vaccines that exist for prevention. Therefore, it is crucial to comprehend the immune response of salmonid species against viruses to develop proactive measures for prevention and control.
Fish have a complex immune system that enables them to defend against pathogenic microorganisms and maintain their overall health [6]. This is achieved through various immune defense mechanisms such as innate immunity and adaptive immunity [7, 8]. The innate immune system relies on germ-line encoded pattern recognition receptors to identify non-self and danger signals, detecting pathogen-associated molecular patterns like bacterial and fungal glycoproteins, lipopolysaccharides, and intracellular components released during injury or infection [9]. The adaptive immune system in fish comprises specialized defense mechanisms that enable them to recognize and mount specific responses against pathogens [10]. These encompass cytokines, T-cell receptors, immunoglobulins, and major histocompatibility complex components [11]. Cytokines, which are small soluble proteins and potent signaling molecules, play a vital role in conveying instructions and facilitating communication among immune and non-immune cells [12]. The cytokine network, essential for hematopoiesis, inflammation, and adaptive immunity, influences a wide range of biological processes, including embryonic development, disease pathogenesis, responses to infection, cognitive function alterations, and the progression of aging-related degenerative processes [13, 14]. Cytokines, such as growth factors, interferons (IFN), tumor necrosis factor-α (TNF-α), interleukins (ILs), and chemokines, are essential for creating a potent antiviral environment within cells and can be categorized into distinct functional classes, including lymphocyte growth factors, pro-inflammatory or anti-inflammatory molecules, and regulators that polarize the immune response to antigens [15]. ILs are a subset of cytokines that regulate intercellular communication within the immune system [16, 17]. Moreover, interleukin-18 (IL-18), a subfamily within the IL-1 cytokine family, is a powerful inflammatory cytokine that triggers the inflammation process [18]. So, to fully harness the potential of fish immunity, a comprehensive understanding of the function of their members, genes, and gene family becomes imperative [19].
In rainbow trout, various gene families play a crucial role in regulating both biotic and abiotic factors. In response to viral infection, they produce IL-1 proteins to initiate and modulate immune reactions. IL-18 gene subfamily members in rainbow trout have been shown to be differentially regulated in response to various stimuli. For example, the expression of IL-18 and its receptors has been found to be upregulated in rainbow trout after infection with bacterial and viral pathogens [20]. In addition, the IL-18 gene family in rainbow trout may have potential as biomarkers for assessing the fish's health and immune status. So, it is important to further investigate the specific functions and mechanisms of the IL-18 gene family in rainbow trout to fully understand their role in immune defense and disease resistance in this species. This study characterizes the IL-18 family genes within the O. mykiss genome to gain insight into these genes' positions, evolutions and functions and evaluate the results through in silico analysis.
MATERIALS AND METHODS
Genome-wide identification of IL-18 gene sequences in O. mykiss
The genomic sequence and annotation details for rainbow trout were obtained from the NCBI genome database. To identify IL-18 genes in O. mykiss, the IL-18 gene sequence of Gallus gallus (XP_046788219.1) was retrieved from the NCBI database for further analysis. Subsequently, the obtained Pfam [21] domain PF00340 sequence was used to discern IL-18 genes in O. mykiss, employing BLAST-P [22] in TBtool with an e-value threshold of ≤1e-5. Physiochemical properties, including the number of amino acids, molecular weight, isoelectric points (pI), and grand average hydrophilicity (GRAVY) values of extracted proteins, were determined using the online tool ExPASy [23]. Utilizing WoLF PSORT [24], predictions for the subcellular localization of IL-18 genes were made. WoLF PSORT incorporates sorting signals, amino acid composition, and functional motifs, such as DNA-binding motifs, to convert protein amino acid sequences into numerical features related to localization.
Phylogenetic analysis
A comparative phylogenetic tree was constructed using MEGA11 software [25] by aligning the IL-18 protein sequences of rainbow Trout (O. mykiss) with the previously reported IL-18 protein sequences of two other fish species, namely Atlantic salmon (Salmo salar) and Goldfish (Carassius auratus). The phylogenetic tree was constructed utilizing the neighbor-joining (NJ) algorithm, and the strength of the groups was evaluated based on bootstrap values obtained from 1000 replications. Then, the phylogenetic tree of IL-18 genes was further refined and enriched through the utilization of the web-based software iTOL [26].
Domain, motif, and gene structure analysis
The identification of conserved domains within each IL-18 gene was conducted using the NCBI CDD tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) [27] and TBtool. Analyzing conserved protein motifs of IL-18 family proteins was performed with the Multiple Em for Motif Elicitation (MEME) program (http://meme-suite.org/tools/meme) [28]. MEME was configured with parameters specifying a maximum motif number of 10 and an optimal motif width ranging from 6 to 100. The examination and illustration of the exon-intron structure of IL-18 genes were achieved using the Gene Structure Display Structure (GSDS) tool [29] and TBtool.
Chromosomal locations and synteny analysis
The rainbow trout genome data from NCBI was used to visualize the chromosomal locations of IL-18 genes using the TBtools program [30]. TBtools was also utilized to create a syntenic map, which revealed the linkages of synteny between orthologous genes in Atlantic salmon and Goldfish, as well as paralogous genes in rainbow trout.
Gene ontology analysis
GO annotations were utilized to conduct an enrichment analysis, assessing the functions of IL-18 genes in rainbow trout. The molecular, cellular, and biological processes of these genes were obtained from the ShinyGo v0.741 web tool [31], available at (http://bioinformatics.sdstate.edu/go/), to perform the GO word enrichment analysis.
Expression analysis of IL-18 genes in liver
To examine the ways in which the IL-18 genes are expressed in rainbow trout liver in response to infectious hematopoietic necrosis virus (IHNV) infection were downloaded from the online NCBI GEO database (Accession: GSE206411) [32]. The Statistix 8.1 pairwise comparison tool was used to identify differences in gene expression between conditions that were statistically significant.
RESULTS
Genome-wide identification of IL-18 gene family in O. mykiss
The study identified a total of nine genes that belong to the IL-18 family, and these genes were named OmIL18-1, OmIL18-2, OmIL18-3, OmIL18-4, OmIL18-5, OmIL18-6, OmIL18-7, OmIL18-8, and OmIL18-9. Comprehensive information of these nine IL-18 genes is provided, including their gene name, gene ID, protein length, chromosomal localization, molecular weights, isoelectric points, instability index, and other pertinent features (Table 1). The analysis revealed proteins with different lengths, ranging from 223 to 822 amino acids, and molecular weights ranging from 25,430.98 to 87,494 kilodaltons. Among the genes, OmIL18-8 has the longest amino acid length (822 amino acids) and the biggest molecular weight (87494 kDa). On the other hand, OmIL18-5 has the lowest amino acid length (223 amino acids) and the smallest molecular weight (25430.98 kDa). These proteins were located on different chromosomes (5, 6, 7, 14, 18, 29, and 32) in the whole genome. OmIL18-1 reported an instability index value below 40, indicating protein stability. However, the other proteins revealed values above 40, indicating their instability. The hydropathy values (GRAVY) indicated that all the genes were primarily hydrophilic. The proteins displayed isoelectric point (pI) values ranging from 5.49 to 7.31, suggesting that they would have a neutral charge within this pH range.
The subcellular localization of genes is crucial for studying diverse aspects of cellular function and biological processes. Among the nine genes examined in this work, OmIL18-3 showed notable expressions in both the cytoplasm and nucleus (Cyto_nucl), as well as in the extracellular space (Extr). OmIL18-5 also displayed notable expressions, mostly in the mitochondria (Mito) and peroxisomes (Pero). OmIL18-2 demonstrated substantial expression in many organelles, including the cytoplasm, nucleus, extracellular space, lysosomes (Lyso), endoplasmic reticulum (E.R.), and peroxisomes. Conversely, OmIL18-1, OmIL18-4, OmIL18-6, OmIL18-7, OmIL18-8, and OmIL18-9 displayed reduced or minor levels of expression in all organelles, with minimal to nonexistent presence in any of these cellular compartments. (Figure 1).
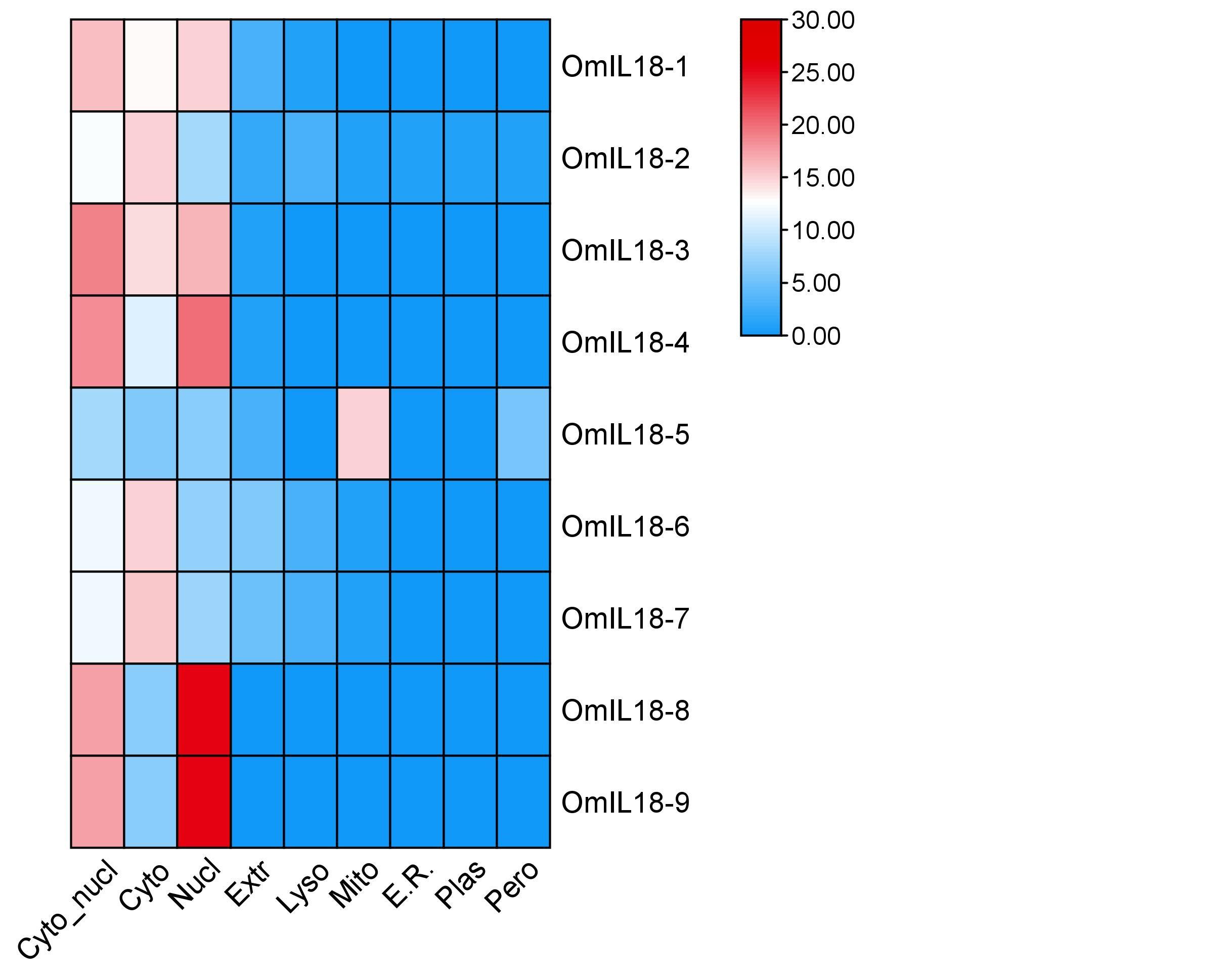
Table 1. Comprehensive overview of IL-18 gene family in rainbow trout (O. mykiss).
Comparative phylogenetic analysis of the IL-18 gene family
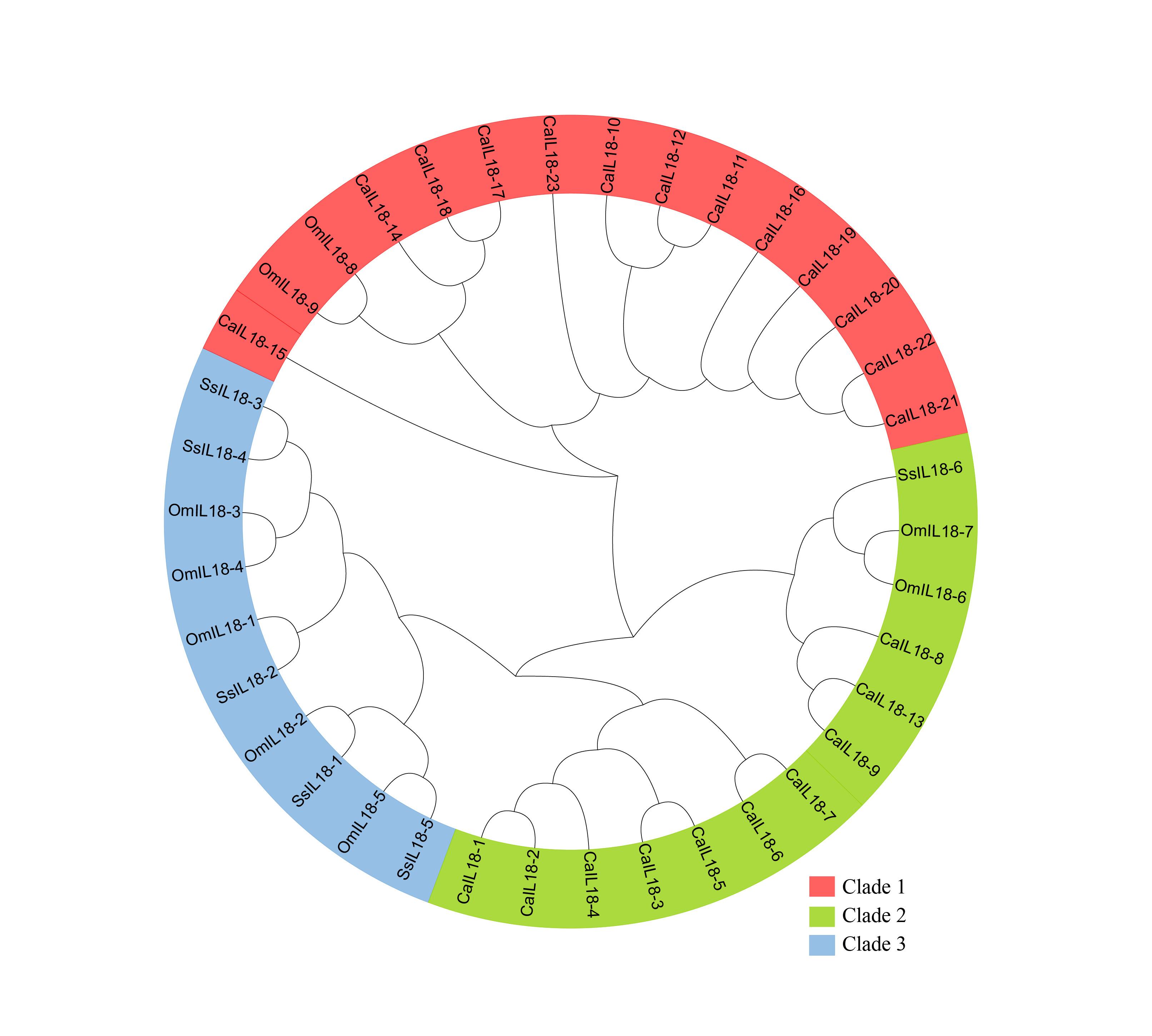
A comparative phylogenetic tree was created to investigate the evolutionary relationships among IL-18 proteins in the three specified species. The phylogenetic analysis categorized the IL18 genes from three fish species into three subgroups, referred to as subfamilies I-III. The analysis encompassed a total of 38 IL18 genes, consisting of 9 from O. mykiss, 23 from Carassius auratus, and 6 from Salmo salar. In the generated phylogenetic tree, each clade grouped together genes with similar functions, and the formation of these clades was based on the characteristics of Salmo salar (Figure 2).
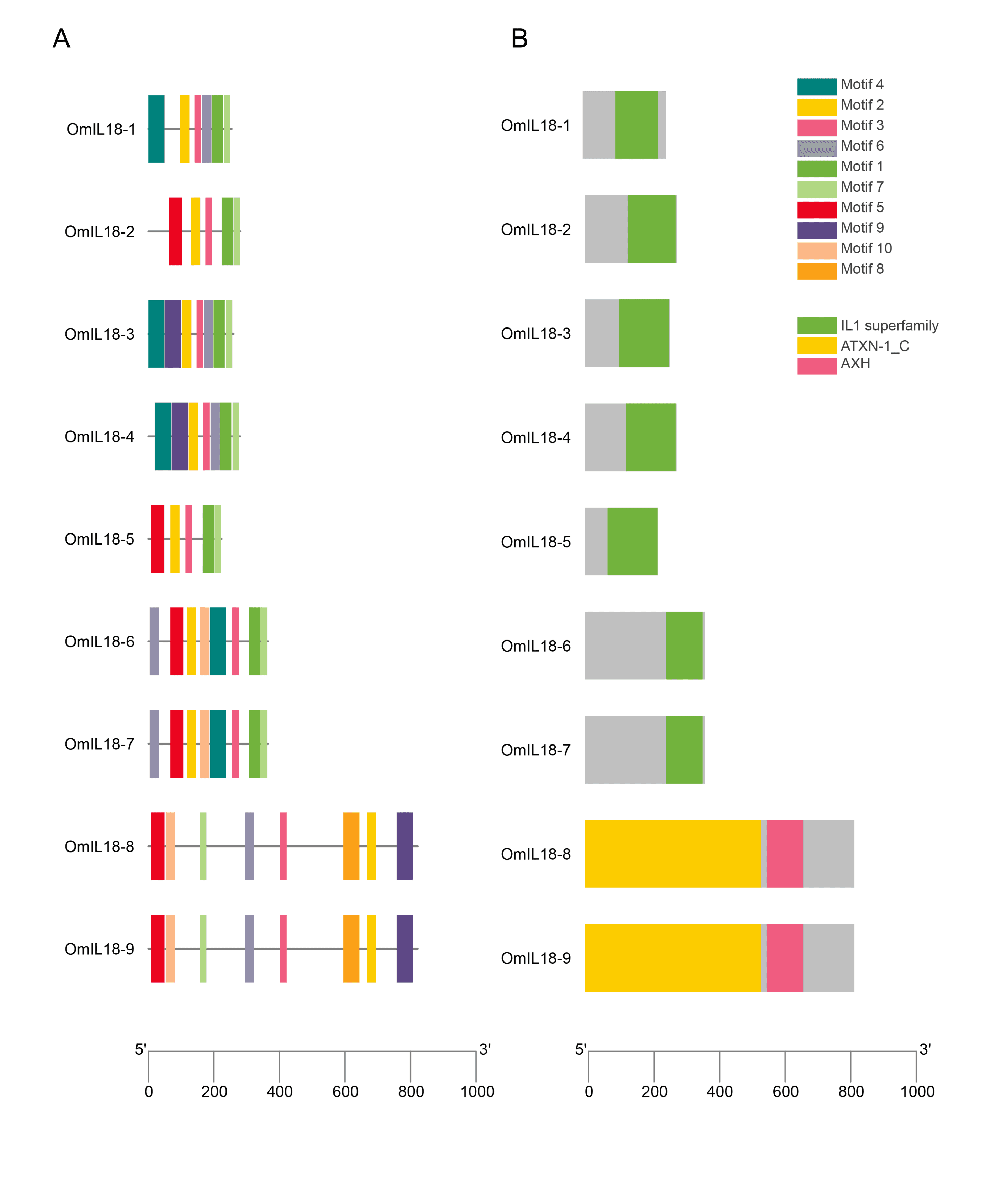
Conserved domain, motif and structure analysis of IL-18 gene family
In all OmIL18 proteins, the IL-18 (IL-1 superfamily) domain was consistently present and conserved. The IL-18 domain was identified as part of the superfamily ATXN-1-C, where AXH serves as a subfamily within the IL-18 superfamily. The motifs within these proteins varied in length, ranging from a maximum of 8 to a minimum of 3. Significantly, among these motifs, motif 3 and motif 2 displayed a high degree of conservation. In addition, IL-18 genes belonging to the same group exhibited motifs with similar patterns, indicating that these conserved motifs are distinctive features of particular groups or subgroups. Although all rainbow trout fish OmIL18 genes shared the same IL18 domain, the arrangement of this domain varied slightly in some of the genes (Figure 3).
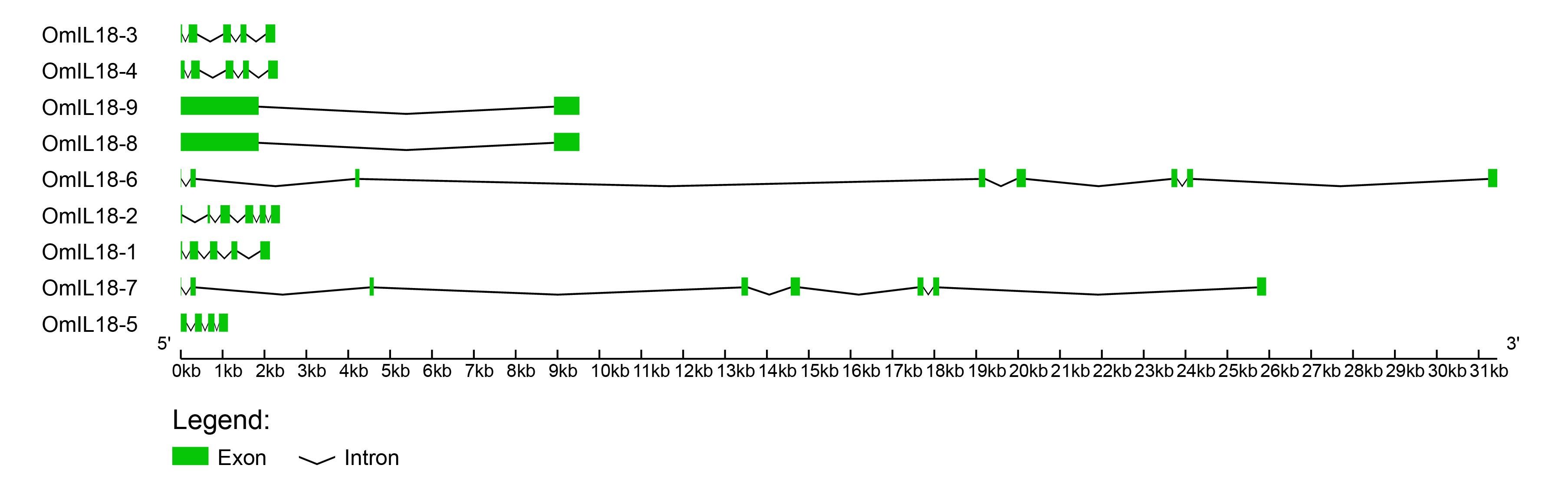
The gene structural properties of IL-18 were examined by analyzing the intron, exon, 5′ UTR, and 3′ UTR structures using the Gene Structure Display Server (GSDS). The number of exons for IL-18 varied between 1 and 7. All OmIL18 genes display a combination of introns and exons, with slight variations in their quantities. For example, OmIL18-9 and OmIL18-8 have the lowest number, consisting of one intron and two exons. Conversely, OmIL18-1, OmIL18-3, and OmIL18-4 each contain four introns and four exons. On the other hand, OmIL18-6 and OmIL18-7 possess the highest number of introns, with 6 in each case, and the most exons, totaling 7. This diversity in the number of introns, ranging from a minimum of 1 to a maximum of 6, and the number of exons, varying from 4 to 7, is a distinctive characteristic of OmIL18 genes (Figure 4).
Chromosomal locations and synteny analysis of IL-18 gene family
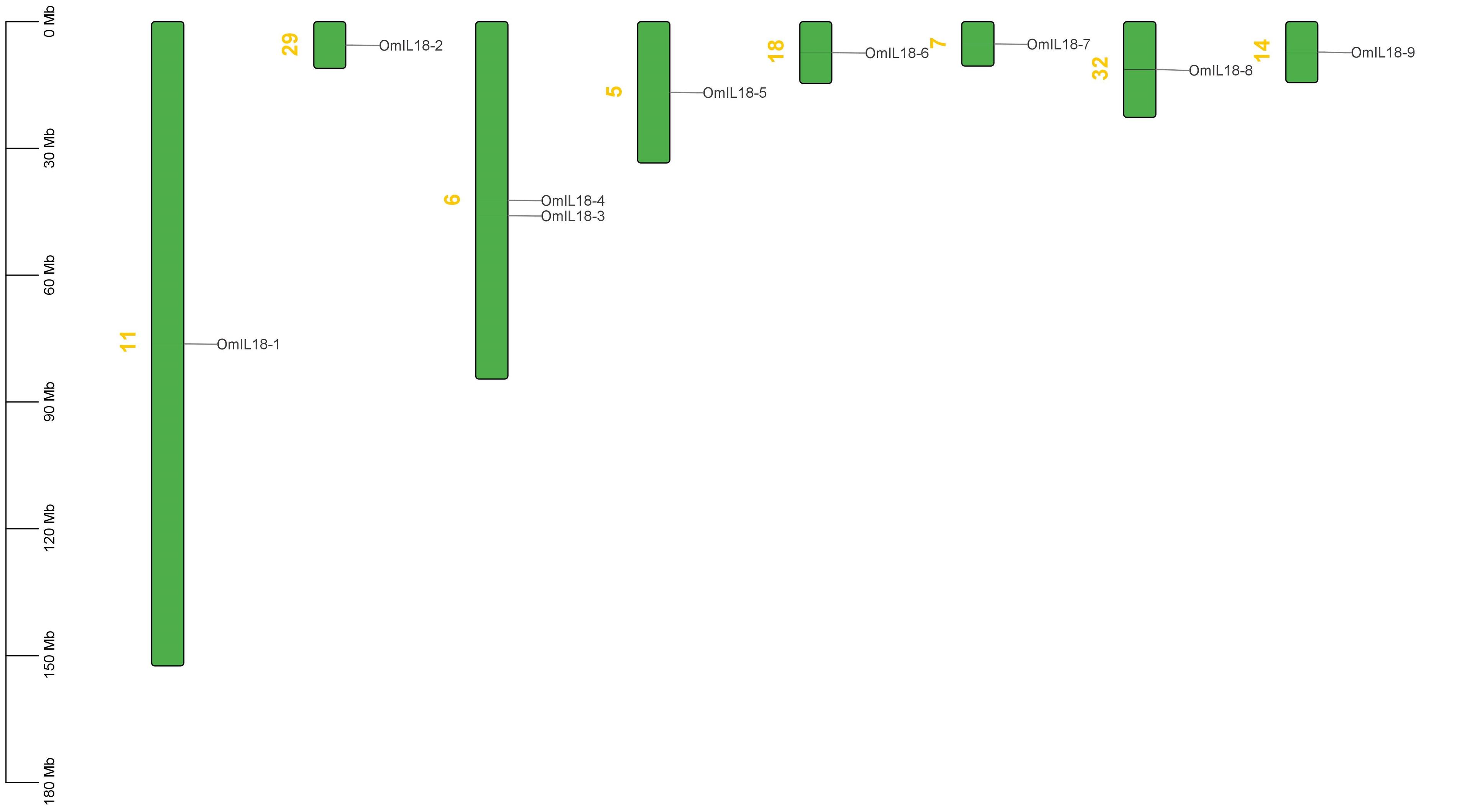
The analysis of chromosomal locations uncovered an uneven distribution of nine candidate IL-18 genes spread across eight chromosomes (5, 6, 7, 11, 14, 18, 28, and 32 Chr). The OmIL18 genes were distributed across different chromosomes: OmIL18-1 on chromosome 11, OmIL18-2 on chromosome 29, OmIL18-4 and OmIL18-3 on chromosome 6, OmIL18-5 on chromosome 5, OmIL18-6 on chromosome 18, OmIL18-7 on chromosome 7, OmIL18-8 on chromosome 32, and OmIL18-9 on chromosome 14 (Figure 5). This analysis provides insights into the spatial arrangement and potential interactions among OmIL18 genes within the rainbow trout fish genome.
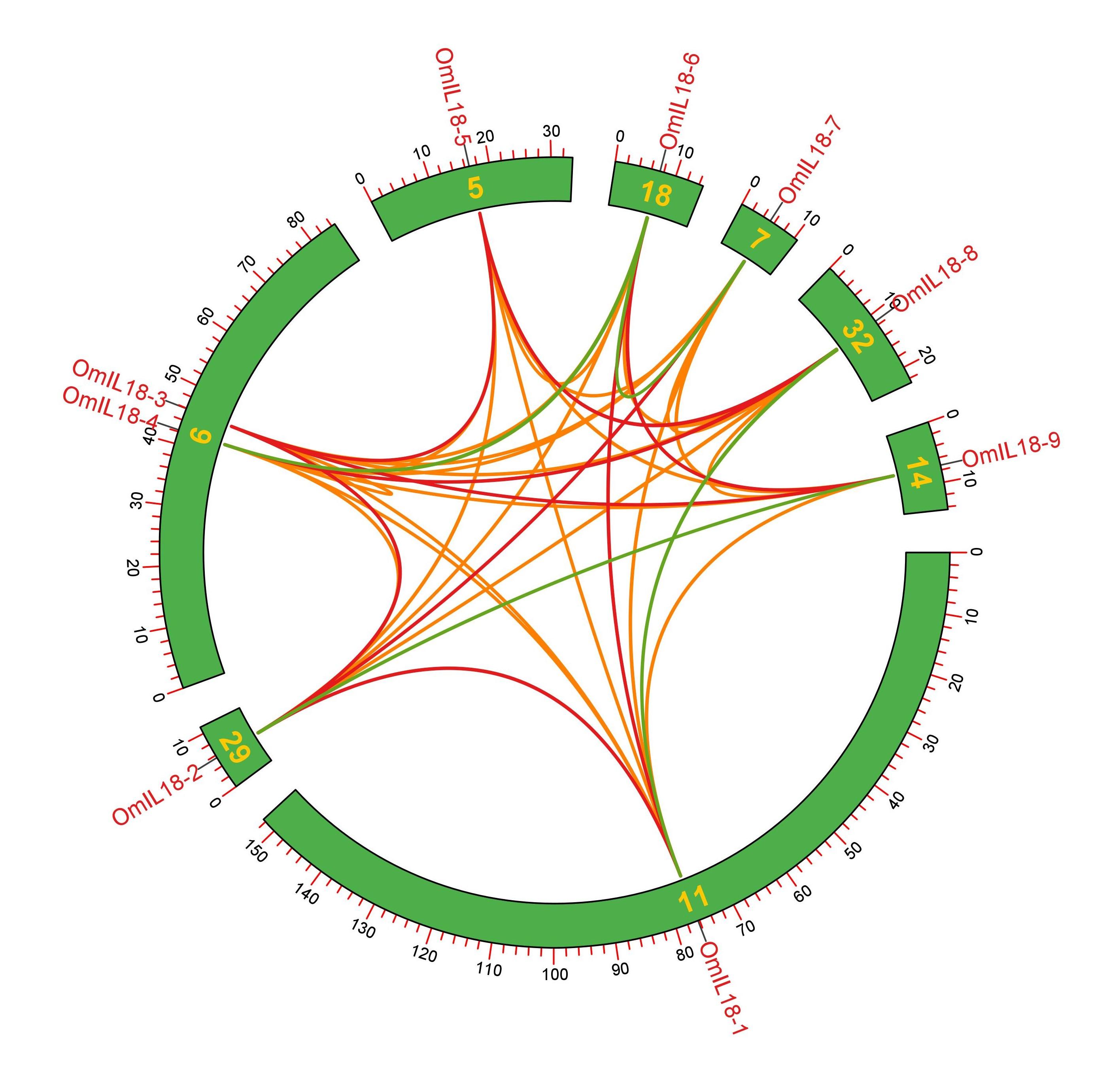
Synteny analysis has revealed the occurrence of both tandem and segmental duplications in the rainbow trout fish genome. For instance, OmIL18-3 and OmIL18-4 underwent tandem duplication on chromosome 6, while OmIL18-5 experienced duplication on chromosome 5. OmIL18-2 was involved in duplication on chromosome 29, OmIL18-8 on chromosome 32, OmIL18-6 on chromosome 18, OmIL18-7 on chromosome 7, OmIL18-9 on chromosome 14, and OmIL18-1 on chromosome 11. These tandem and segmental duplications are significant in the context of evolution, contributing to genetic diversity, gene family expansion, and potential functional innovations in the rainbow trout fish genome (Figure 6).
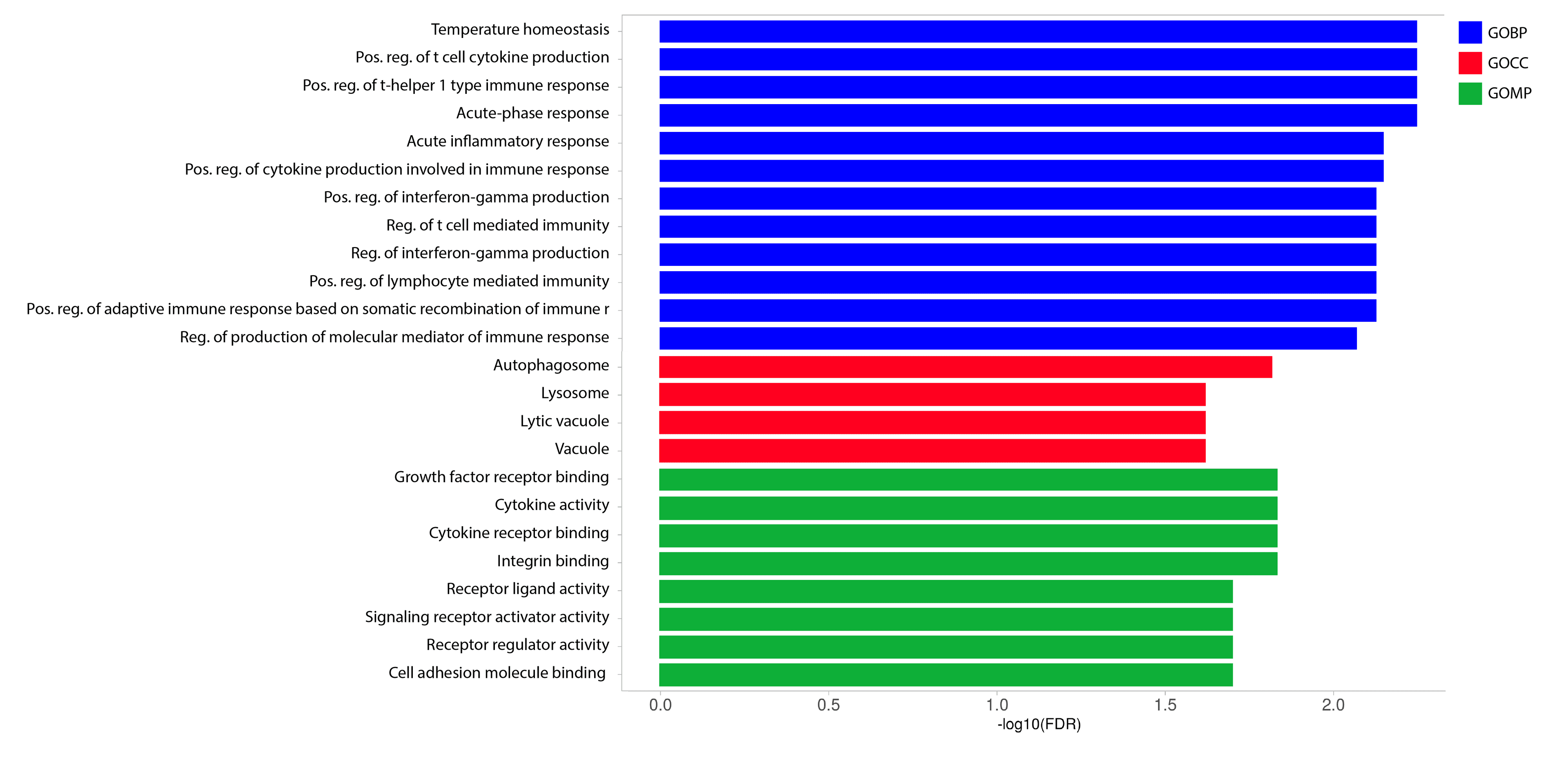
Gene ontology (GO) analysis of IL-18 gene family
The fold enrichment analysis of the OmIL18 gene family revealed three distinct functional categories: molecular, biological, and cellular functions. Among these, the molecular functions that stood out included growth receptor binding (GO:0005102) and cytokine activity (GO:0005125). In the analysis of OmIL18 genes, notable biological functions included temperature homeostasis (GO:0001659) and cell cytokine production (GO:0001816). Additionally, a key cellular function observed was autophagy (GO:0005776) (Figure 7).
Expression analysis of IL-18 gene in the liver in response to hematopoietic necrosis virus
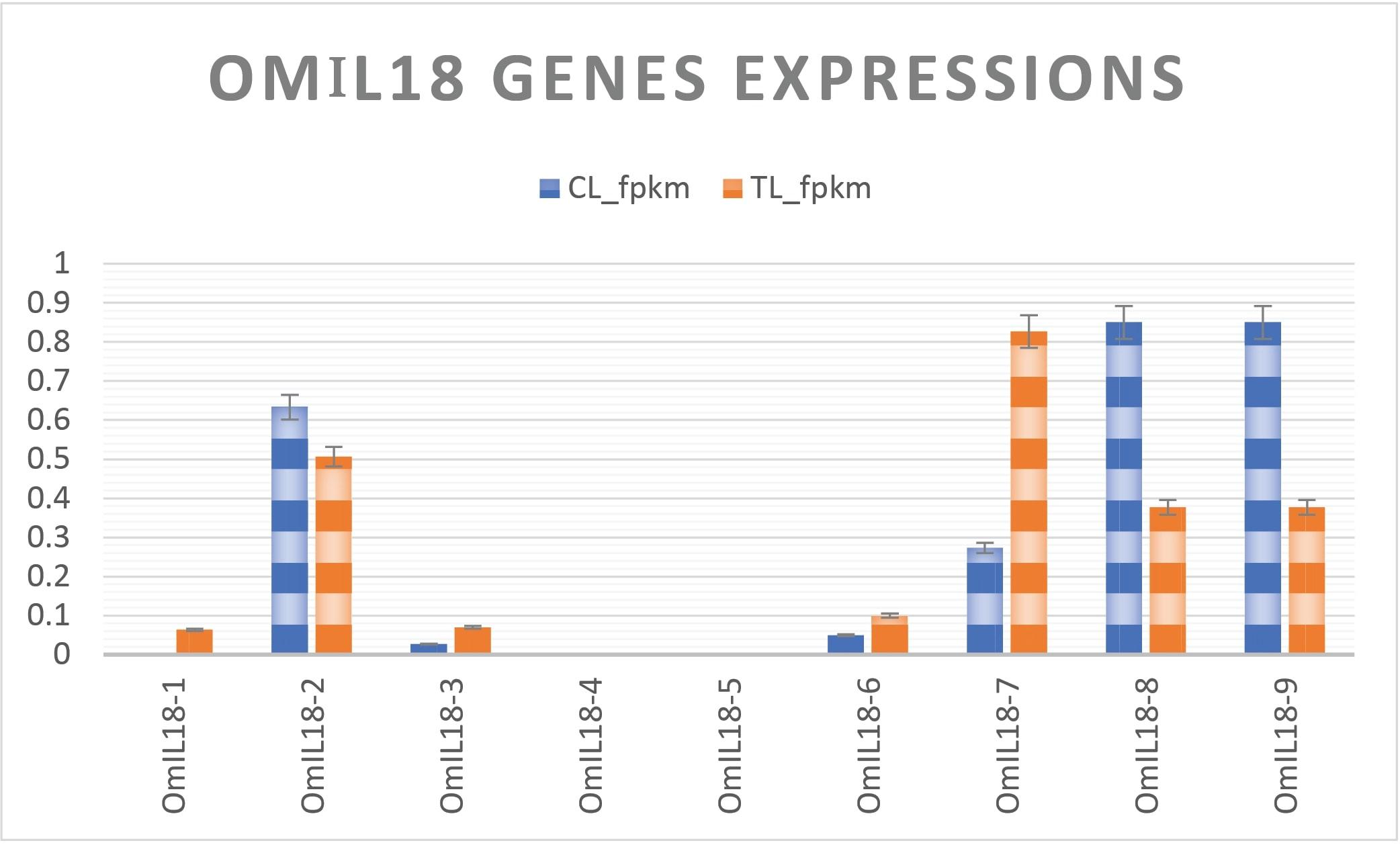
RNA-Seq data on the expression of immune genes in the liver of rainbow trout were retrieved from the NCBI Geo database under Accession: GSE206411, following infection with the infectious hematopoietic necrosis virus [20]. The FPKM (fragments per kilobase per million reads) values were computed based on the RNA-seq data downloaded from the database. In the liver of rainbow trout afflicted with the infectious hematopoietic necrosis virus, the analysis of gene expression revealed diverse responses.
OmIL18-1 exhibited significantly elevated expression in the treatment group, whereas OmIL18-2, OmIL18-8, and OmIL18-9 were downregulated. OmIL18-7 displayed a substantial and highly significant upregulation, while OmIL18-3 showed a modest increase. Both OmIL18-4 and OmIL18-5 consistently exhibited low expression levels in both groups. Overall, during the attack of the hematopoietic necrosis virus, OmIL18-1, OmIL18-3, OmIL18-6, and OmIL18-7 were upregulated. These alterations in gene expression portray the dynamic response of the liver of rainbow trout to the viral infection (Figure 8).
DISCUSSION
In this study, a comprehensive genome-wide investigation was conducted to identify and characterize IL-18 genes within the rainbow trout genome. The study of the physicochemical properties of IL-18 gene encoded proteins is essential for predicting their behavior, designing experiments, and engineering proteins for specific purposes in fields like medicine, biotechnology, and biochemistry. The OmIL18 genes reveal common hydrophilic characteristics indicated by negative GRAVY values. Additionally, the instability index highlighted those eight proteins that displayed characteristics of instability, while one showed notable stability. The subcellular localization analysis revealed a wide range of organelle distribution, with a substantial proportion mainly found in the cytoplasm, nucleus, and extracellular space.
Phylogenetic analysis allows us to trace the evolutionary relationships between these genes and understand how they have adapted and diversified in response to different ecological and environmental factors. In this study, the identification of distinct subgroups (subfamilies I-III) in the comparative phylogenetic tree indicates the diversification of these genes over time, offering valuable insights into the mechanisms of gene evolution and their contributions to the diversity and adaptability of these fish species. The interesting clade formation based on Salmo salar traits suggests that these genes, which came from a common ancestor, have gone on separate evolutionary paths in each species. In earlier, several studies have validated that the immune genes in O. mykiss have evolved differently in each species, with evidence of divergent selection and adaptive evolution [33, 34].
The presence of the IL18 domain in all OmIL18 proteins is essential for their functionality, particularly in immune system processes and inflammatory responses. This domain is found in many proteins, which suggests that they all play similar roles in immune system functions. Highly conserved motifs like motif 3 and motif 2 are very important for keeping essential protein functions going. The similar motif patterns within genes of the same group imply shared functional roles or evolutionary histories among these genes. Moreover, the variation in intron-exon structures among OmIL18 genes underscores their evolutionary history, indicating that certain genes may have evolved for specific functions while others have preserved ancestral features. This diversity contributes to the genetic complexity and adaptability of rainbow trout fish. Previous studies found that antiviral immunity in mammals relies on the diverse mechanisms employed by TRIM proteins [35]. The main innate immune signaling pathways, such as Toll-like receptor and IFN signaling, are conserved in mammals and teleost fish [36]. The evolution and conservation of the IFN system, which plays a crucial role in the antiviral innate immune response in both teleosts and mammals [37].
In order to gain insights into the spatial arrangement and dispersion of IL-18 genes across the genome, we systematically mapped them onto their corresponding chromosomes. This comprehensive visualization provides a detailed representation of the specific locations of IL-18 genes within the chromosomal framework, allowing for a more thorough understanding of their genomic distribution and potential implications. Chromosomal mapping helps in understanding the evolution of whole genomes [38]. Besides, chromosomal mapping provides crucial insights into how the genome has changed, enabling the study of the evolution of complex traits and regulatory networks [39]. The investigation into the distribution of OmIL18 genes across various chromosomes in the rainbow trout fish genome highlights the impact of gene duplication, loss, and organization on evolutionary processes. Additionally, this distribution can influence gene regulation, potentially leading to the co-regulation of genes located on the same chromosome, thereby influencing their functions and expression patterns. Genes on the same chromosome may share a common evolutionary history, while those on different chromosomes have diversified over time. This information is crucial for studying genome evolution, including the evolution of complex traits and regulatory networks [40]. Fish are thought to harbor a greater number of genes compared to other vertebrates, potentially impacting a genome duplication event that occurred during the evolution of the teleost fish lineage [41]. The glutamine synthetase genes in O. mykiss are organized into separate linkage groups, which could mean that they share functional roles [42]. Furthermore, a study identified 14 immune system-related genes in rainbow trout, highlighting their clustering in distinct genomic blocks, which remain conserved across various fish species despite independent evolution and instances of genome duplication [43]. Synteny analysis of this study plays a crucial role in deciphering these duplication events, as it helps identify conserved gene order and evolutionary relationships among duplicated genes. Synteny analysis revealing both tandem and segmental duplications in the rainbow trout fish genome is highly relevant to understanding the evolutionary dynamics of this species [44]. These duplication events suggest a complex history of gene evolution and adaptation. Tandem duplications can lead to the expansion of specific gene families, potentially offering the fish new genetic resources for coping with environmental changes or evolving novel functions [45]. Segmental duplications, which involve larger DNA segments, can contribute to genetic diversity and the formation of distinct genetic subpopulations [46]. The expansion of the opsin gene family in fish is predominantly facilitated by tandem duplication, revealing a distinct correlation between opsin gene repertoires and variations in spectral environment, morphological traits, or life history traits [47].
The gene ontology analysis of the OmIL18 gene family demonstrates their significant participation in several biological processes, including growth receptor binding and cytokine activity, which have an impact on signal transduction and immunological responses. Biologically, these processes play important roles in maintaining a stable internal temperature, producing cell cytokines, and regulating cellular function for autophagy. They are significant for maintaining internal balance, controlling immune-related processes, and affecting gene expression in response to environmental changes and immune challenges.
Gene expression patterns of IL-18 gene family members in rainbow trout could provide insights into the immune response mechanisms and help in developing novel strategies for improving disease resistance and immune modulation in rainbow trout aquaculture. Notably, OmIL18-1, OmIL18-7, and, to some extent, OmIL18-3 and OmIL18-6 showed upregulation, indicating their potential roles in the immune response against the virus. Upregulated genes are often associated with immune defense mechanisms, such as the production of antiviral proteins or signaling molecules [20]. In earlier studies, substantial activation was noted in key signaling pathways, such as the Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, apoptosis, RIG-I-like receptor signaling pathway, p53 signaling, and JAK-STAT signaling pathway, in response to IHNV infection [48, 49]. In addition, the presence of IHNV infection caused the activation of specific genes, such as Mx-1, viral hemorrhagic septicemia virus-induced gene 8 (Vig-8), TNF-α1, TNF-α2, IL-1β1, IL-8, and Hsp70 [50, 51]. Moreover, scientists observed a swifter and more pronounced up-regulation of cytokine genes at elevated water temperatures. When subjected to vaccination with a Yersinia ruckeri bacterin at a high temperature of 25°C, a significant increase in the expression of proinflammatory cytokines, namely IL-1β1 and INF-γ, was noted [52]. Besides viral infection, bacterial and parasitic infections induced varied expressions of immune genes in rainbow trout [53]. Upon immersion exposure to Yersinia ruckeri or intraperitoneal injection with Flavobacterium psychrophilum, increased levels of IL-8, INF-γ, TNF-α, and IgM gene expressions were also observed [54]. Similarly, elevated expressions of immunoregulatory genes such as IL-1 beta isoform 1 (IL-1b1), TNF-α isoforms 1 and 2 (TNF- α1 and TNF- α2), and IL-8 genes were noted in amoebic gill disease in rainbow trout [55]. Moreover, significant up-regulation of IL-1β and IL-8 was observed in symptomatic fish in response to L. garvieae bacterial infection [56]. Trout skin infected with Gyrodactylus derjavini parasite exhibited heightened expression of TNF-α1, TNF-α2, TGF-beta, and IL-8 genes [57].
This investigation suggests that the IL-18 gene family in rainbow trout could have a crucial role in bolstering the immune response against pathogens. Exploring the evolutionary patterns, expression dynamics, and regulation of IL-18 gene family members in rainbow trout holds promise for uncovering immune response mechanisms. Such insights could prove instrumental in devising innovative strategies to enhance disease resistance and modulate immunity in rainbow trout aquaculture. However, it's important to note that while genome-wide studies can anticipate gene functions, the necessity for functional validation through experimental methods remains imperative to validate the anticipated roles. Additionally, it's acknowledged that the study may not encompass the complete range of tissue-specific or developmental stage-specific expression patterns within the IL-18 gene family.
CONCLUSION
This thorough analysis of the entire genome provides valuable insights into the essential function of the IL-18 gene family in rainbow trout. This study offers vital insights into the genetic and physiological aspects of these genes by examining their evolutionary patterns, physicochemical properties, subcellular localization, and interactions. The IL-18 gene family's evolutionary and functional dynamics are better understood through the identification of discrete subgroups, the conservation of domains and motifs, and the heterogeneous distribution of genes across chromosomes. Moreover, the study highlights the significance of these genes in immunological reactions, specifically in relation to disease resistance and regulation in aquaculture. To fully use the genes' capacity to improve disease resistance and immunity in rainbow trout aquaculture, it is imperative to do additional study and experimental verification.
ACKNOWLEDGEMENT
None.
AUTHORS CONTRIBUTION
SMB designed outlines and drafted the manuscript. SMB and MIH performed the experiments and analyzed the data. MRI and MMUH reviewed the scientific contents described in the manuscript. All authors read and approved the final submitted version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]D’Agaro E, Gibertoni P, et al. Recent trends and economic aspects in the rainbow trout (Oncorhynchus mykiss) sector. Appl Sci. 2022; 12: 8773.
- [2]Duman M, Altun S, et al. A review of bacterial disease outbreaks in rainbow trout (Oncorhynchus mykiss) reported from 2010 to 2022. J Fish Dis. 2023; 00: 1–17.
- [3]Khalil SM, Orioles M, et al. Current knowledge of lactococcosis in rainbow trout: Pathogenesis, immune response and prevention tools. Aquaculture. 2024; 580: 740363.
- [4]Collet B. Innate immune responses of salmonid fish to viral infections. Dev Comp Immunol. 2014; 43(2): 160-73.
- [5]He M, Ding NZ, et al. Novirhabdoviruses versus fish innate immunity: A review. Virus Res. 2021; 304: 198525.
- [6]Mokhtar DM, Zaccone G, et al. Main components of fish immunity: An overview of the fish immune system. Fishes. 2023; 8(2): 93.
- [7]Uribe C, Folch H, et al. Innate and adaptive immunity in teleost fish: a review. Vet Med (Praha). 2011; 56(10): 486-503.
- [8]Sahoo S, Banu H, et al. Immune system of fish: an evolutionary perspective. Antimicrobial Immune Response. IntechOpen: UK, 2021, pp 1-21.
- [9]Magnadottir B. Innate immunity of fish (overview). Fish Shellfish Immunol. 2006; 20(2): 137-51.
- [10]Kordon AO, Pinchuk L, et al. Adaptive immune system in fish. Turkish J Fish Aquat Sci. 2021; 22(4): TRJFAS20235
- [11]Warr GW. The adaptive immune system of fish. Dev Biol Stand. 1997; 90: 15-21.
- [12]Arango Duque G, Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 2014; 5: 491.
- [13]Zou J, Secombes CJ. The function of fish cytokines. Biology. 2016; 5(2): 23.
- [14]Dinarello CA. Historical insights into cytokines. Eur J Immunol. 2007; 37: S34-S45.
- [15]Savan R, Sakai M. Genomics of fish cytokines. Comp. Biochem. Physiol. Part D Genomics Proteomics. 2006; 1: 89-101.
- [16]Secombes CJ, Wang T, et al. The interleukins of fish. Dev Comp Immunol. 2011; 35(12): 1336-45.
- [17]Kaiser P, Rothwell L, et al. Evolution of the interleukins. Dev Comp Immunol. 2004; 28(5): 375-394.
- [18]Dinarello CA, Novick D, et al. Interleukin-18 and IL-18 binding protein. Front Immunol. 2013; 4: 289.
- [19]Abo-Al-Ela HG. An introduction to selected innate immune-relevant genes in fish. Appl Ecol Environ Res. 2018; 16(2): 955-976.
- [20]Wu S, Huang J, et al. Integrated analysis of immune parameters, miRNA-mRNA interaction, and immune genes expression in the liver of rainbow trout following infectious hematopoietic necrosis virus infection. Front Immunol. 2022; 13: 970321.
- [21]Sonnhammer EL, Eddy SR, et al. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998; 26(1): 320-322.
- [22]Eric SD, Nicholas TD, et al. Bioinformatics with basic local alignment search tool (BLAST) and fast alignment (FASTA). JBSA. 2014; 6(1): 1-6.
- [23]Duvaud S, Gabella C, et al. Expasy, the swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021; 49: W216-W27.
- [24]Horton P, Park KJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007; 35(2): W585-W587.
- [25]Tamura K, Stecher G, et al. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021; 38(7): 3022-3027.
- [26]Letunic I, Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021; 49(W1): W293-W296.
- [27]Marchler-Bauer A, Zheng C, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2012; 41: D348-D352.
- [28]Bailey TL, Johnson J, et al. The MEME suite. Nucleic Acids Res. 2015; 43(W1): W39-W49.
- [29]Hu B, Jin J, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015; 31(8): 1296-1297.
- [30]Chen C, Chen H, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020; 13(8): 1194-1202.
- [31]Ge SX, Jung D, et al. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020; 36(8): 2628-2629.
- [32]Barrett T, Suzek TO, et al. NCBI GEO: mining millions of expression profiles-database and tools. Nucleic Acids Res. 2005; 33(1): D562-D566.
- [33]Limborg MT, Blankenship SM, et al. Signatures of natural selection among lineages and habitats in Oncorhynchus mykiss. Ecol Evol. 2012; 2: 1-8.
- [34]Ortutay C, Vihinen M. Conserved and quickly evolving immunome genes have different evolutionary paths. Hum Mutat. 2012; 33(10): 1456-1463.
- [35]Langevin C, Levraud JP, et al. Fish antiviral tripartite motif (TRIM) proteins. Fish Shellfish Immunol. 2019; 86: 724-733.
- [36]Stein C, Caccamo M, et al. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 2007; 8: 1-23.
- [37]Langevin C, Aleksejeva E, et al. The antiviral innate immune response in fish: evolution and conservation of the IFN system. J Mol Biol. 2013; 425(24): 4904-4920.
- [38]Cheng Y, Shang D, et al. Whole genome-wide chromosome fusion and new gene birth in the Monopterus albus genome. Cell Biosci. 2020; 10: 1-4.
- [39]Voldoire E, Brunet F, et al. Expansion by whole genome duplication and evolution of the sox gene family in teleost fish. PLoS One. 2017; 12(7): e0180936.
- [40]Van Hellemont R, et al. Divergence of regulatory sequences in duplicated fish genes. InGene and Protein Evolution. Karger Publishers. 2007, pp. 81-100.
- [41]Kim MS, Seo JS, et al. Duplication of phospholipase C-δ gene family in fish genomes. Genomics. 2008; 92(5): 366-71.
- [42]Gharbi K, Murray BW, et al. Genome organization of glutamine synthetase genes in rainbow trout (Oncorhynchus mykiss). Cytogenet Genome Res. 2007; 116(1-2): 113-5.
- [43]Phillips RB, Ventura AB, et al. Mapping rainbow trout immune genes involved in inflammation reveals conserved blocks of immune genes in teleosts. Anim Genet. 2013; 44: 107-13.
- [44]Berthelot C, Brunet F, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014; 5: 3657.
- [45]Shaw CH, Gao G, et al. Differential expression and evolution of three tandem, interleukin-1 receptor-like 1 genes in rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol. 2018; 87: 193-203.
- [46]Soylev A, Le TM, et al. Discovery of tandem and interspersed segmental duplications using high-throughput sequencing. Bioinformatics. 2019; 35(20): 3923-3930.
- [47]Rennison DJ, Owens GL, et al. Opsin gene duplication and divergence in ray-finned fish. Mol Phylogenet Evol. 2012; 62(3): 986-1008.
- [48]Pan Y, Huang J, et al. Dynamic immune response in the spleens of rainbow trout (Oncorhynchus mykiss) to infectious hematopoietic necrosis virus revealed by transcriptome and immune-related genes expression analysis. Aquac Rep. 2023; 29: 101473.
- [49]Zhao L, Huang J, Li Y, Wu S, Kang Y. Comprehensive analysis of immune parameters, mRNA and miRNA profiles, and immune genes expression in the gill of rainbow trout infected with infectious hematopoietic necrosis virus (IHNV). Fish Shellfish Immunol. 2023; 133: 108546.
- [50]Purcell MK, Kurath G, et al. Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish Shellfish Immunol. 2004; 17(5): 447-462.
- [51]Tafalla C, Coll J, et al. Expression of genes related to the early immune response in rainbow trout (Oncorhynchus mykiss) after viral haemorrhagic septicemia virus (VHSV) infection. Dev Comp Immunol. 2005; 29(7): 615-626.
- [52]Raida MK, Buchmann K. Temperature-dependent expression of immune-relevant genes in rainbow trout following Yersinia ruckeri vaccination. Dis Aquat Org. 2007; 77(1): 41-52.
- [53]Khalil SM, Bulfon C, et al. Immune profiling of rainbow trout (Oncorhynchus mykiss) exposed to Lactococcus garvieae: Evidence in asymptomatic versus symptomatic or vaccinated fish. J Fish Dis. 2023; 46(7): 731-41.
- [54]Evenhuis JP, Cleveland BM. Modulation of rainbow trout (Oncorhynchus mykiss) intestinal immune gene expression following bacterial challenge. Vet Immunol Immunopathol. 2012; 146(1): 8-17.
- [55]Bridle AR, Morrison RN, et al. The expression of immune-regulatory genes in rainbow trout, Oncorhynchus mykiss, during amoebic gill disease (AGD). Fish Shellfish Immunol. 2006; 20(3): 346-364.
- [56]Khalil SM, Sacca E, et al. In field study on immune-genes expression during a lactococcosis outbreak in rainbow trout (Oncorhynchus mykiss). Aquaculture. 2023; 574: 739633.
- [57]Lindenstrom T, Secombes CJ, et al. Expression of immune response genes in rainbow trout skin induced by Gyrodactylus derjavini infections. Vet Immunol Immunopathol. 2004; 97(3-4): 137-48.