Exploring impact of integrated breeding strategies in enhancing yield, nutritional quality, and stress tolerance in alfalfa
Abstract
Yield, nutrition quality and stress tolerance are important traits for alfalfa improvement perspective. These qualitative and quantitative attributes are changed during its life cycle, no updated studies available on whether and how these traits are influenced by several environmental factors. Therefore, we updated the role of several breeding strategies for developing alfalfa yield, nutritional quality and biotic-abiotic stress tolerance in alfalfa. This study explored integrated breeding approaches would be suitable for the desire traits improvement in alfalfa. Subsequently, the integration of multiomics including genomics, transcriptomics, proteomics, metabolomics, and ionomics may facilitate the agronomic traits improvement and plant fitness in alfalfa. Furthermore, this study proposes integration of omics-system with top-down (phenotype to genotype) and bottom-up (genotype to phenotype) model that can be helpful to characterize or develop desire qualitative and qualitative traits in alfalfa. This updated study might be useful to alfalfa breeders and farmers for improving alfalfa through breeding programs.
INTRODUCTION
Alfalfa (Medicago sativa L.), known as “Queen of the Forages” is an important forage legume. Alfalfa is widely cultivated as a forage crops with good source of protein, and alternative nitrogen source [1]. These positive attributes lead to consider it as ideal crop for livestock and dairy production. Alfalfa is an autotetraploid (2n = 4x = 32), allogamous, perennial legume crop cultivated worldwide [2]. Most of the varieties of cultivated alfalfa originated from subsp. sativa or subsp. × varia, or by its other sub-species hybridization [3]. Forage yield, nutritional quality, and the ability of stress tolerance and/or resistance are key agronomic traits in alfalfa plants. Several qualitative attributes traits such as crude protein (CP), neutral detergent fiber (NDF), acid detergent fiber (ADF), and acid detergent lignin (ADL) are important features for ensuring fodder quality of alfalfa [3]. Unfortunately, these traits are either genetically control or influenced by multiple environmental stress stimuli [4]. Several biotic and abiotic stresses significantly inhibit plant growth, decline forage productivity (Table 1), which lead to decline market demand and utilization of alfalfa fodder in livestock industry [5-7]. Therefore, cost effective breeding strategies are highly demandable.
Breeding approaches are exciting tool for improving of yield, quality and stress tolerance in alfalfa [8]. Several traditions to breeding approves are commonly used for improving alfalfa form the ancient time to current era [3].The traditional breeding approaches provide excellent output for alfalfa improvement, but these approaches are very time consuming, it may not produce desire results in alfalfa [9]. Therefore, a combined breeding programs of traditional, molecular and modern breeding approaches would be exciting, low cost effective and farmers friendly for improving alfalfa yield, nutrition quality, stress tolerance in alfalfa plants. The modern breeding approach based on molecular marker, multiomics information would be suitable for the improvement of traits associated with forage yield and its quality [10, 11]. The farmers traditionally cultivate local germplasms, which are tolerant to multiple stresses and able to adapt arid and semi-arid environments. Unfortunately, some landrace populations are replaced by high yielding cultivars [12]. The uncontrolled introduction of the alfalfa cultivars occurs erosion of genetic diversity (GD), low sustainability to changing environments, which lead to loss of performance (LOP) and productivity of alfalfa [13].
In these above background scenarios, many modern alfalfa breeders are applying multiomics and gene engineering approaches for developing key agronomic traits in alfalfa without losing their performances [4]. Application of modern breeding approaches have made excellent progress in improving alfalfa desire traits from lab to field levels. In this review, we explored the limitations of traditional breeding for improving agricultural and qualitative attributes in alfalfa, and provided update understanding on how the modern breeding strategies solve the limitations of traditional breeding in case of alfalfa plants. Further, we emphasized the impact of integrated breeding strategies in enhancing yield, nutritional quality, and stress tolerance in alfalfa.
ENVIRONMENTAL FACTORS INFLUENCING YIELD, QUALITY, AND FITNESS IN ALFALFA
Factors influencing alfalfa yield
Alfalfa yield productivity negatively regulated by several factors including seed yield breeding cultivars, cropping system, growth, crop durability, dry matter production efficiency [14]. Alfalfa yield can also be influenced by fertilization usage, genetic barrier, water use efficiency (WUE)[15]. The production of alfalfa regulated by cropping system including soil depth, drainage system, fertilization, and deficit irrigation [16]. Though alfalfa supports in suppling water in dry season through deep root system. Water is a critical factor where rooting depth (8-30 feet) can vary the amount of water availability as well as dry matter production. Deep soil provides better alfalfa yield compared to shallow soils. However, deficit irrigation decline biological nitrogen fixation that lead to decline alfalfa yield [17]. Application of nitrate fertilization and intercropping system significantly influences the nodule formation, dry matter yield and resource use efficiency in alfalfa [18]. The above studies imply that several factors associated with alfalfa life cycle, are involved in declining yield in alfalfa (Figure 1).
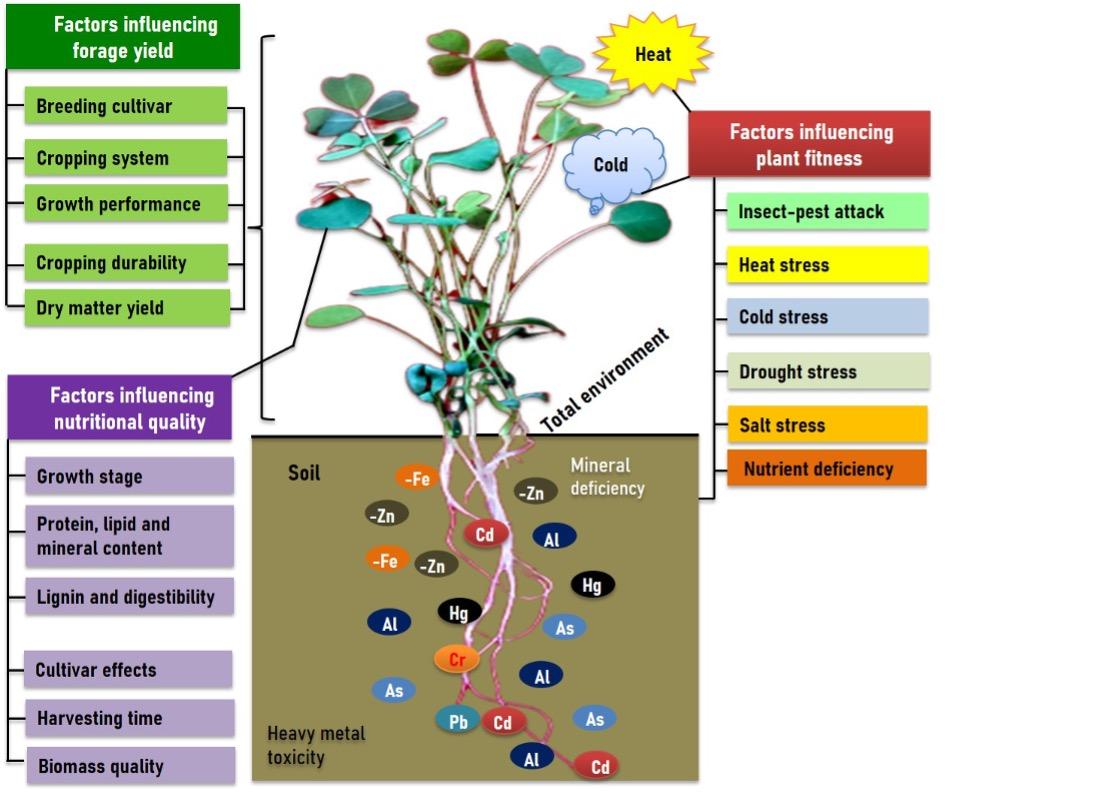
Factors influencing alfalfa nutritional quality
Alfalfa is an excellent resources of livestock animals that considered as sustainable source of food proteins [19]. Its nutritional quality alters during plant growth stage and life cycle. The chlorophyll content, amino acids, and flavonoids levels considerably declines at maturity stage compared to other stages in alfalfa [20]. Lignin is a phenolic polymer linked with cellulose, hemicellulose and pectin in the cell wall, high level of lignin influences feed digestibility in livestock animals [21]. As forage crop, alfalfa nutritional status depends on the concentration of carbohydrates, proteins, lipids and present of other phenolic compounds. The arrangement of these organic nutrients decides the digestibility of each crop, along with mineral and mineral values (D-value) and sum of energy that can be obtained by an animal [21].The accessibility of degrading enzymes to cellulose is restricted by high lignification levels in secondary cell walls; thus, lignin downregulation by genetic modification was used to improve the quality of the alfalfa [22]. Modification of lignin content in alfalfa has been followed in previous studies using both conventional plant breeding methods and transformation methods through lignin-lowering enzymes [22, 23]. The plant harvesting time influences the nutritional quality and silage of alfalfa [24]. Therefore, it is clear that several factors associated with alfalfa growth, developments are directly or partially involved in regulating nutritional attributes in alfalfa (Figure 1).
Impact of biotic and abiotic factors in alfalfa
Several biotic and abiotic factors including insect pests attack, heat, cold, drought, soil salinity, temperatures, nutrient deficiency are involved in declining growth, productivity and nutrition quality in alfalfa. In the following sections, we stated how abiotic and abiotic stress regulates growth, productivity and plant fitness in alfalfa. Insect pests attract under changing climate are critical factors inhibits normal growth, development and plant productivity in the world [25]. The chlorophyll (a, b) content, soluble proteins and dry matter yield significantly declined due to increase of number of thrips attack in alfalfa [26]. This study indicates that interactions among the crops, climate and insects is crucial for damaging crop and yield loss. In this scenario, insect pests can cause more crop damage and productivity loss due to changing climate.
In contrast, multiple abiotic stresses are drastically declined alfalfa growth, yield and nutritional quality in alfalfa (Figure 1). The differences of growing condition significantly influence alfalfa productivity. In a recent study on total 197 alfalfa cultivars suggest that temperature higher than 25°C (>25°C) limits alfalfa growth, and biomass yield [27], while cold stress (<10°C) causes growth retardation, necrosis, decline photosynthetic rare and significand yield in alfalfa [1]. Alfalfa exhibits water scarcity slowly due to deep root system compared to other legumes, but the growth, productivity and protein content considerably declined in response to drought stress in alfalfa [28]. However, stress induced oxidative stress causes cellular injury and even plant death. The high level of salt stress severely causes leaf chlorosis, low photosynthesis performance, decline forage biomass yield and its quality. Previous study reveals that excess sodium toxicity declined plant growth performance, and altered proteomic changes in alfalfa [11].
Mineral deficiency is another key limitation of alfalfa that lead to decline plant growth, fitness, forage biomass yield and its quality (Figure 1). Recently, growth and physiological impairments, and reduction of Zn, S and Ca concentration found in Fe-starved alfalfa [29]. Furthermore, multiple heavy metals (HMs) toxicity severely inhibit plant growth, development and productivity. For instance, the alfalfa growth, biomass yield, mineral nutrition found to be impaired in response to toxic pollutants [6]. Mercury toxicity considerably causes nutrient (Fe, S) reduction in alfalfa [5], while cadmium toxicity causes plant retardation, chlorosis, low photosynthetic performance and biomass yield in alfalfa [30]. Further, Hg toxicity induces oxidative stress in alfalfa that lead to inhibit plants normal growth, photosynthesis and ultimate biomass yield in alfalfa [31]. Therefore, it is clear that mineral deficiency stress along with HMs toxicity directly involved in declining growth, yield and nutritional quality in alfalfa, such type of scenario is presented in case of alfalfa (Figure 1).
REVISITING THE ROLES OF TRADITIONAL BREEDING AND MODERN BREEDING APPROACHES IN IMPROVING ALFALFA TRAITS
Traditional breeding approach
Alfalfa is a autotetraploid (2n = 4x = 32) perenial species. Improvment of alfalfa production using treaditional breeding is a long term approach, along with several limitations including tetrasomic inheritance, inbreeding depression, and genotype-environment interaction are always challenging. Despite of these, treaditional breeding approcah has significant importance for improving desireble traits in alfalfa [32].
Table 1. Impacts of biotic-abiotic stress on alfalfa forage yield and its nutritional quality.
Forage quality has been improved using alfalfa breeding. Milic et al. [41], explored genetically control of important forage quality related traits in leaves and stems of alfalfa genotypes, They found the highest values of narrow sense heritability for NDF and CP in leaves, while ADF was most heretable traits in stems, suggested that nutritive value can be improved using alfalfa breeding program [41]. Study suggestes that recurrent phynotypic selection would be effective for improving yield, and tolerence to biotic and abiotic stresses in alfalfa [42]. However, molecular breeding along with other modern breeding approaches have been implemented lately. However, many disease resistance, and pest management, and high yielding cultivars have been developed using these breeding strategies.
Molecular breeding come modern breeding approaches
The significance of QTLs Mapping or gene discovery, marker assisted section (MAS) and genetic engineering or genetic transformation are emphaised in this section (Figure 2). Alfalfa traits improvement using treaditional breeding is time consuming and costly.
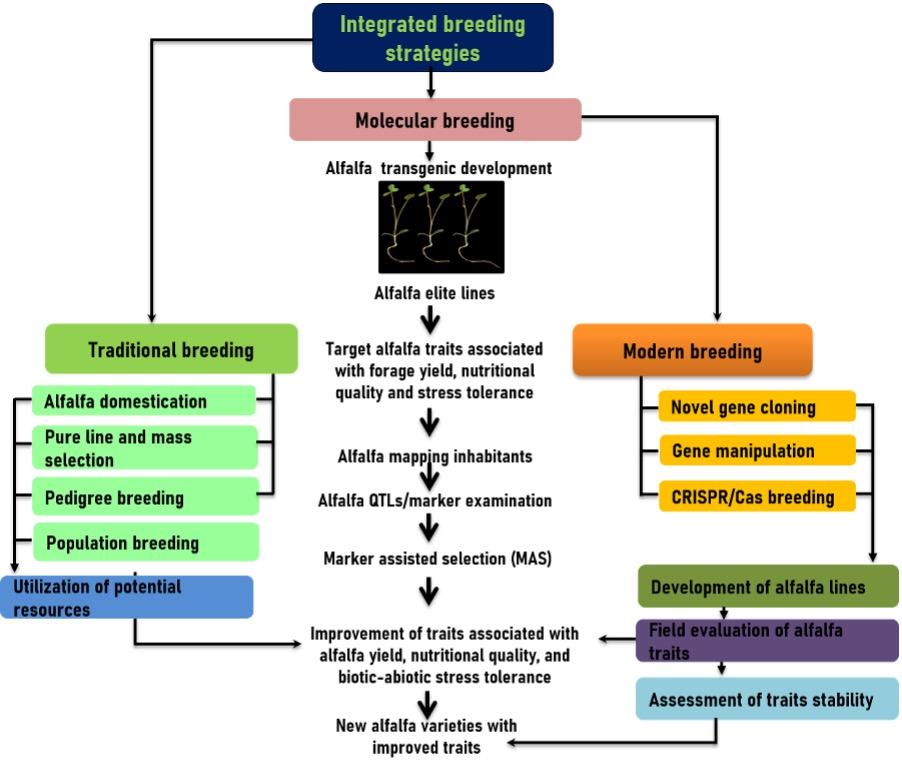
Therefore incorporated MAS in to alfalfalfa breeding programs provide an alternative for alfalfa traits improvement. Thought MAS has been minorly applied for commercial improvement of alfalfa compared to other plant species [43]. Identification and development of genetic loci associated with stress tolerance is the initial step of stress tolerant alfalfa variety using MAS. The Quantitative trait loci (QTL) mapping and genome-wide association studies (GWAS) are key approaches for identifying QLTs, which regulate the several traits including yield, forage quality and stress tolerance in plants including alfalfa [43]. The involvement of QTLs associated with forage yield, nutritional quality and stress tolerance in alfalfa (Table 2). Single-nucleotide polymorphism (SNPs) in a region of the genome with high linkage disequilibrium. Total 198 alfalfa accessions were used to SNPs-associated drought stress tolerance by association mapping (AM). Several functional key genes such as MsHSP23, MsZIP, MsZEP, MsMYB2L, and MsWRKY11 associaed with drought stress tolerance were identificed in alfalfa [8, 43].
In developing modern plant breeding era, gene engineering tool extensively applied in improving insect pests resistance, biotic-abiotic stress resistancein alfafla (Table 3). These breeding staregies lead to producation of novel compounds, improving plant yield, enhancing of indrustrial and/or pharmaceutical proteins in alfalfa [44, 45]. In modern breeding age, the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 approach is being reapidely used for improving alfalfa yield, nutritional quality and stress tolernce in alfalfa plants [46, 47]. The RNAi-mediated down-regulation of alfalfa SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 8 (MsSPL8) was involved in increasing plant brancing, enhanced biomass yield, regrowth and drought toleance in alfalfa [47]. However, despite of the above nutritional and agronomic benefits alfalfa posseses some limitation in usage as pasture and hay forage. A risk factor is initiated specially for tumenents due to high degration of forage protein. More importantly, high lignin content induces the degradation of carbohydrate in rumen as alfalfa shows an unbalanced and asynchronous degradation of nitrogen to carbohydrate (N/CHO) ration in rumen. In this context, gene engineering tool used for improving nutritive value, yeid and performence of stress tolerance in alfalfa [48].
MULTI-OMICS APPROACHES FOR IMPROVING YIELD, QUALITY AND STRESS TOLERANCE IN ALFALFA
High quality of alfalfa is an excellent resource for livestock animals and altimate global food security. Alfalfa plant fitness, nutritional quality, and ptoductivity dramatically change during its life cycle. The multiomics approch drawn great attention for developing qualitative and quantitative traits in alfalfa (Figure 3). In this section, we updated recent advances and future prospects of multiomic approach in improving alfalfa yield, nutritional quality and biotic and abiotic stress tolerance. A progress of genomics and genomic selection in alfalfa has been stated [10]. Recently, Liu et al. [20] reveal new insights into nutritainal quality alteration in alfalfa leaves during flowering time using multi-omics approaches. They emphasized correlation and network analyses based on leaf metabolome, transcriptome and proteome data, which revealed chlorophyll, flavanoid and amino acid content in alfalfa [20].Two contrusting alfalfa genotypes have been considered to characterization of salt stress responsive protein using a proteome approach [11]. Alfalfa leagume tolerates drought stress due to deep root system. A series of drough stress responsive proteins has been identified using proteomic approach, which were involved in physilogical alteration and drough stress stress toleance in alfalfa [28]. A combined transcriptome and proteome appaoch has been used for discovering of key factors involved in water loggning stress in alfalfa [49], where total 16,899 differentially expressed genes (DEGs) were identified. The abscisic acid and gibberellin-related genes regulation patterns at alfalfa develpmental stages analysed by transcriptome approach [50]. These candidate genes were involved in hormone regulation pathway and seed development that opens a new avenue to improve seed yield in alfalfa. Recently, 220 alfalfa cultivars sequences and conduced a genome-wide association (GWA) study using 875023 SNPs [51], while 1,924, 2,405, and 3,779 differentially expressed genes (DEGs) identified at three different growth stages in alfalfa. The GWA combined with transcriptome analyses reveal that genetic loci and key genes lead the flowering time in alfalfa. The series of candidate genes have great significance to breeders for alfalfa traits improvement using breeding programme. However, proper breeding strategies are highly required for desirable traits improvement in alfalfa. Medina et al. [52], yield and vigor traits in response to salt stress in alfalfa, where genotype-by-sequencing (GBS) based marker with allele dose and phenotypic data analysed by GWA. The GWA identified 27 SNPs associated with salt stress tolerance in autotetraploid alfalfa (2n = 4x = 32). The GWAS was applied for identifying mapping loci associated with alfalfa growth and forage yield under salt stress [53].
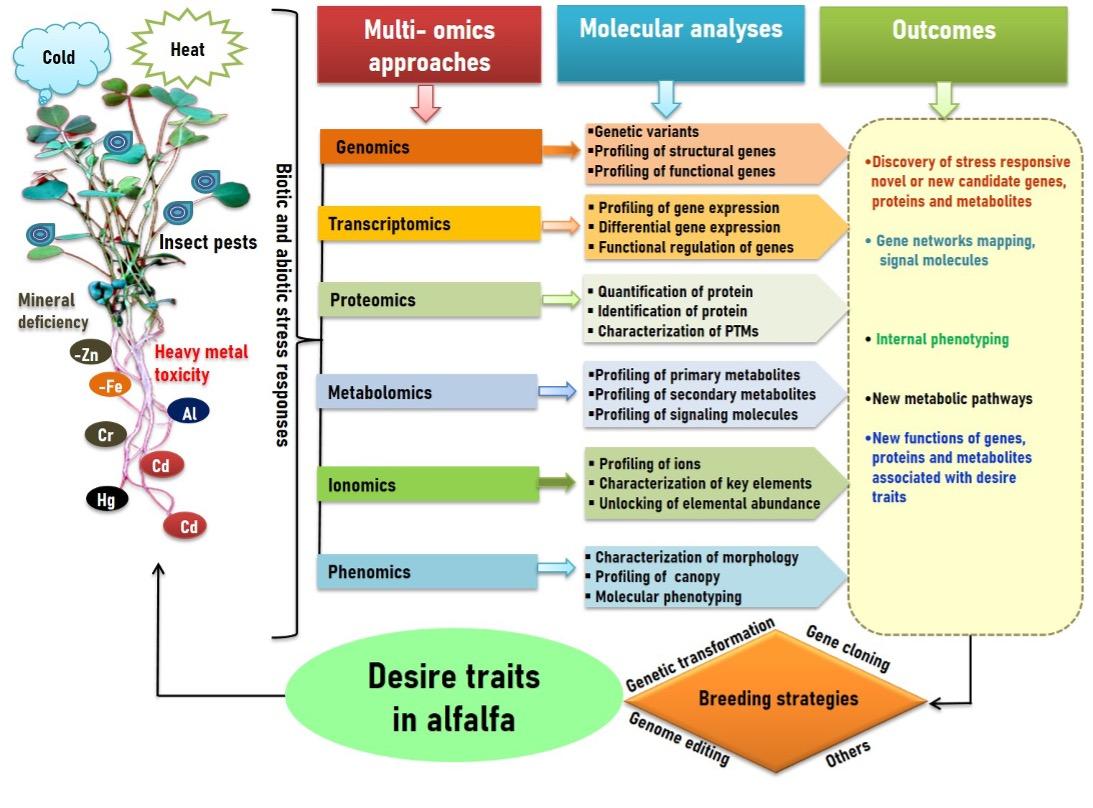
Plant phynotyping plays a key role in alfalfa breeding, along with contribute to explore the link between genotypes and phynotypes. Phenomics-assisted selection approch applied for herbage accumulation in alfalfa [54], where remote sensing technology has been frequintly using in alfalfa for herbage accumulation needs for phynotyping effors such as screeing of large breeding population [55]. The applciation of ionome plays in screening, characterization and distribution of elements in a system. Advances in ionome in alfalfa studies is still elusive. In a study earlier, roots and shoots of seven alfalfa populations screened to examine nickel (Ni) binding ability from aqueous solution, where reported that 80% of Ni bound to the plant tissue and the Ni abundance was 4.1 mg g-1 plant biomass. In contrast, alfalfa was able to remove Ni by 90% from tissues [56]. Isonomic studies have not been well studies in alfalfa. Therefore, this approach can be applied for ions identification, characterization in alfalfa. The above discussions imply the multiomics-tools could be effective for improving yield, nutritional quality and stress tolerance in alfalfa.
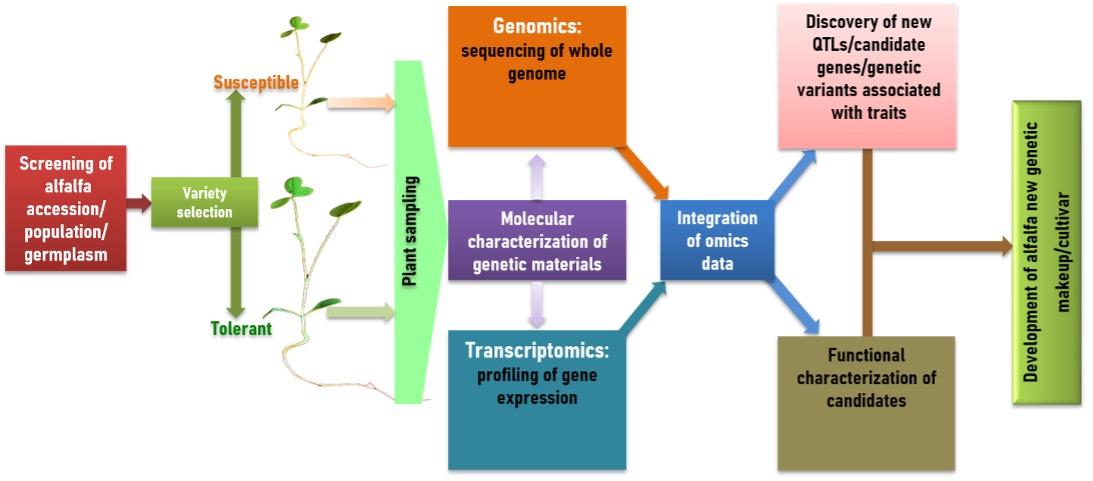
The multiomics tool can also be used for new elite genotype development of alfalfa (Figure 4). In this contrast, two contrasting genotype can be considered for addressing population sampling and subsequent molecular analysis including gene and genome sequencing, gene expression profiling, omics data integration, QTLs identification, functional analyses and ultimately screen and/development of new elite plant genotypes with desire traits (Figure 4).
Table 2. QTLs involved in yield, nutritional quality, and biotic-abiotic stress tolerance in alfalfa
Table 3. Improvement of yield, quality, and biotic-abiotic stress tolerance in alfalfa through molecular breeding
CONCLUSIONS AND FUTURE DIRECTIONS
Improvement of qualitative and quantitative traits in alfalfa plants is critical for its overall improvement. This study provides undated insights into the impact of integrated breeding strategies in enhancing yield, nutritional quality, and stress tolerance in alfalfa. We have described significance of molecular breeding including gene engineering or manipulation approaches for improving new traits and plant fitness in alfalfa plants. Additionally, the potential role of integrated traditional breeding and modern breeding approaches in improving yield, nutritional value and biotic-abiotic stress tolerance in alfalfa. Though such breeding strategies has already been implemented in several alfalfa species but there is still scope to improve alfalfa traits using multiomics molecular tools. Furthermore, the integration of panomics tool combined with multiomics approaches (genomics, transcriptomics, proteomics, metabolomics, phenomics, and ionomics) would facilitate to identify novel/new candidate genes, proteins, phenotypes and metabolites to develop desire traits and plant fitness in alfalfa. The above advances will be useful to alfalfa breeders and farmers for developing new alfalfa cultivars with improved yield, nutritional quality and stress tolerance.
ACKNOWLEDGEMENT
The authors would like to express their gratitude to ABEx Bio-Research Center, Bangladesh for inspiration, and eager engagement to this work.
AUTHORS CONTRIBUTION
AKA and AM conceived the research plan. AKA wrote initial draft. SKB, AK revised and edited the manuscript. All the authors approved the final version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Rahman MA, Song Y, et al. Mechanistic basis of silicon mediated cold stress tolerance in alfalfa (Medicago sativa L.). Silicon. 2023.
- [2]Živković B, Radović J, et al. Assessment of genetic diversity among alfalfa (Medicago sativa L.) genotypes by morphometry, seed storage proteins and RAPD analysis. Ind Crops Prod. 2012;40:285-91.
- [3]Kulkarni KP, Tayade R, et al. Harnessing the potential of forage legumes, alfalfa, soybean, and cowpea for sustainable agriculture and global food security. Front Plant Sci. 2018;9:1314.
- [4]Hrbáčková M, Dvořák P, et al. Biotechnological perspectives of omics and genetic engineering methods in alfalfa. Front Plant Sci. 2020;11:592.
- [5]El-Shehawi AM, Rahman MA, et al. Mercury toxicity causes iron and sulfur deficiencies along with oxidative injuries in alfalfa (Medicago sativa). Plant Biosyst. 2022;156:284-91.
- [6]Mbarki S, Talbi O, et al. Comparison of grain sorghum and alfalfa for providing heavy metal remediation of sandy soil with different soil amendments and salt stress. Front Environ Sci. 2022;10.
- [7]Lee K-W, Rahman MA, et al. Identification of cold and heat-induced differentially expressed genes in alfalfa (Medicago sativa L.) Leaves. JAPS. 2017;27: 1231-1237.
- [8]Lee K-W, Rahman M, et al. Expression of small heat shock protein23 enhanced heat stress tolerance in transgenic alfalfa plants. JAPS. 2017;27:1238-44.
- [9]Shi S, Nan L, et al. The current status, problems, and prospects of alfalfa (Medicago sativa L.) breeding in China. Agronomy. 2017;7:1.
- [10]Hawkins C, Yu L-X. Recent progress in alfalfa (Medicago sativa L.) genomics and genomic selection. Crop J. 2018;6:565-75.
- [11]Rahman MA, Alam I, et al. Screening for salt-responsive proteins in two contrasting alfalfa cultivars using a comparative proteome approach. Plant Physiol Biochem. 2015;89:112-22.
- [12]Rhouma HB, Taski-Ajdukovic K, et al. Assessment of the genetic variation in alfalfa genotypes using SRAP markers for breeding purposes . J Chil.J.Agric.Res. 2017;77:332-9.
- [13]Wang H, Coulman B, et al. Genetic diversity and local adaption of alfalfa populations (Medicago sativa L.) under long-term grazing. Sci Rep. 2023;13:1632.
- [14]Tucak M, Horvat D, et al. Assessment of alfalfa populations for forage productivity and seed yield potential under a multi-year field trial. Agronomy2023.
- [15]Li Y, Su D. Alfalfa water use and yield under different sprinkler irrigation regimes in north arid regions of China. Sustainability2017.
- [16]Liu M, Wang Z, et al. Effect of regulated deficit irrigation on alfalfa performance under two irrigation systems in the inland arid area of midwestern China. Agric Water Manag. 2021;248:106764.
- [17]Kamran M, Yan Z, et al. Irrigation and nitrogen fertilization influence on alfalfa yield, nutritive value, and resource use efficiency in an arid environment. Field Crops Res. 2022;284:108587.
- [18]Zhao Y, Liu X, et al. Effect of root interaction on nodulation and nitrogen fixation ability of alfalfa in the simulated alfalfa/triticale intercropping in pots. Sci Rep. 2020;10:4269.
- [19]Hadidi M, Orellana Palacios JC, et al. Alfalfa as a sustainable source of plant-based food proteins. Trends Food Sci Technol 2023;135:202-14.
- [20]Liu Y, Fan W, et al. Multi-omics analyses reveal new insights into nutritional quality changes of alfalfa leaves during the flowering period. Frontiers in plant science. 2022;13:995031.
- [21]Zhong H, Zhou J, et al. The effect of lignin composition on ruminal fiber fractions degradation from different roughage sources in water buffalo (Bubalus bubalis). 2021;Agriculture, 11:1015.
- [22]Barros J, Temple S, et al. Development and commercialization of reduced lignin alfalfa. Curr Opin Biotechnol. 2019;56:48-54.
- [23]Reddy MSS, Chen F, et al. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). 2005;PNAS,102:16573-8.
- [24]Liu X, Li D, et al. Effects of harvest period and mixed ratio on the characteristic and quality of mixed silage of alfalfa and maize. Anim Feed Sci Technol 2023;306:115796.
- [25]Subedi B, Poudel A, et al. The impact of climate change on insect pest biology and ecology: Implications for pest management strategies, crop production, and food security. J Agric Food Res. 2023;14:100733.
- [26]Wu F, Shi S, et al. Physiological and biochemical response of different resistant alfalfa cultivars against thrips damage. Physiol Mol Biol Plants 2021;27:649-63.
- [27]Kim JY, Han KJ, et al. Assessment of growing condition variables on alfalfa productivity. Animal Sci Technol. 2023;65:939-50.
- [28]Rahman MA, Kim Y-G, et al. Proteome analysis of alfalfa roots in response to water deficit stress. J Integr Agric. 2016;15:1275-85.
- [29]Rahman MA, Ahmed MB, et al. Growth and physiological impairments in Fe-starved alfalfa are associated with the downregulation of Fe and S transporters along with redox imbalance. Chem Biol Technol Agric. 2021;8:36.
- [30]Das U, Rahman MA, et al. Sulfur triggers glutathione and phytochelatin accumulation causing excess Cd bound to the cell wall of roots in alleviating Cd-toxicity in alfalfa. Chemosphere. 2021;262:128361.
- [31]Rahman MA, Kabir AH, et al. Glutathione restores Hg-induced morpho-physiological retardations by inducing phytochelatin and oxidative defense in alfalfa. Biology 2020;9.
- [32]Acharya JP, Lopez Y, et al. Breeding alfalfa (Medicago sativa L.) adapted to subtropical agroecosystems. 2020;Agronomy,10:742.
- [33]Berberet RC, Morrison RD, et al. Impact of the alfalfa weevil, Hypera postica (Gyllenhal) (Coleoptera: Curculionidae), on forage production in nonirrigated alfalfa in the southern plains. J Kans Entomol. 1981;54:312-8.
- [34]Cuperus GW, Radcliffe EB, et al. Economic injury levels and economic thresholds for pea aphid, Acyrthosiphon pisum (Harris), on alfalfa. Crop Prot. 1982;1:453-63.
- [35]P. RV, B. VE, et al. Aphid damage to alfalfa hay. California Agriculture. 1956:pp:1-2.
- [36]Latheef MA, Caddel JL, et al. Alfalfa production as influenced by pest stress and early first harvest in Oklahoma. Crop Prot. 1988;7:190-7.
- [37]Ray IM, Townsend MS, et al. Heritabilities of water-use efficiency traits and correlations with agronomic traits in water-stressed alfalfa. Crop Sci. 1999;39:494-498.
- [38]Peterson PR, Sheaffer CC, et al. Drought effects on perennial forage legume yield and quality. Agron J. 1992;84:774-9.
- [39]Sanz-Sáez Á, Erice G, et al. Alfalfa forage digestibility, quality and yield under future climate change scenarios vary with Sinorhizobium meliloti strain. J Plant Physiol. 2012;169:782-8.
- [40]Saeed IAM, El-Nadi AH. Irrigation effects on the growth, yield, and water use efficiency of alfalfa. Irrig Sci. 1997;17:63-8.
- [41]Milić D, Karagić Đ, et al. Breeding and improvement of quality traits in alfalfa (Medicago sativa ssp sativa L.). 2014;Genetika,46:11-8.
- [42]Biazzi E, Nazzicari N, et al. Genome-wide association mapping and genomic selection for alfalfa (Medicago sativa) forage quality traits. PLOS ONE. 2017;12:e0169234.
- [43]Jiang X, Yu A, et al. Identification of QTL and candidate genes associated with biomass yield and Feed Quality in response to water deficit in alfalfa (Medicago sativa L.) using linkage mapping and RNA-Seq. Front Plant Sci. 2022;13:996672.
- [44]Wang D, Khurshid M, et al. Genetic engineering of alfalfa (Medicago sativa L.). Protein Pept Lett. 2016;23:495-502.
- [45]Kumar T, Bao A-K, et al. The progress of genetic improvement in alfalfa (Medicago sativa L.). Czech J Genet. 2018;54:41-51.
- [46]Bao Q, Wolabu TW, et al. Application of CRISPR/Cas9 technology in forages. Grassland Res.. 2022;1:244-51.
- [47]Singer SD, Burton Hughes K, et al. The CRISPR/Cas9-mediated modulation of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 8 in alfalfa leads to distinct phenotypic outcomes. Front Plant Sci.. 2021;12:774146.
- [48]Lei Y, Hannoufa A, et al. The use of gene modification and advanced molecular structure analyses towards improving alfalfa forage. Int J Mol Sci. 2017;18.
- [49]Zeng N, Yang Z, et al. Comparative transcriptome combined with proteome analyses revealed key factors involved in alfalfa (Medicago sativa) response to waterlogging stress. Int J Mol Sci. 2019;20.
- [50]Zhao L, Li M, et al. Transcriptome analysis and identification of abscisic acid and gibberellin-related genes during seed development of alfalfa (Medicago sativa L.). BMC Genom. 2022;23:651.
- [51]He F, Zhang F, et al. A Genome-wide association study coupled with a transcriptomic analysis reveals the genetic loci and candidate genes governing the flowering time in alfalfa (Medicago sativa L.). Front Plant Sci.. 2022;13:913947.
- [52]Medina CA, Hawkins C, et al. Genome-wide association and prediction of traits related to salt tolerance in autotetraploid alfalfa (Medicago sativa L.). Int J Mol Sci. 2020;21.
- [53]Liu XP, Yu LX. Genome-wide association mapping of loci associated with plant growth and forage production under salt stress in alfalfa (Medicago sativa L.). FrontPlant Sci. 2017;8:853.
- [54]Biswas A, Andrade M, et al. Phenomics-assisted selection for herbage accumulation in alfalfa (Medicago sativa L.). Frontiers in plant science. 2021;12:756768.
- [55]Cazenave A-B, Shah K, et al. High-throughput approaches for phenotyping alfalfa germplasm under abiotic stress in the field. Plant Phenome J. 2019;2:190005.
- [56]Gardea Torresdey J, Tiemann K, et al. Ability of Medicago sativa (alfalfa) to remove nickel ions from aqueous solution. Proceedings of the 10 th Annual Conference on Hazardous Waste Research,2000. p. 239-48.
- [57]Ray IM, Han Y, et al. Identification of quantitative trait loci for alfalfa forage biomass productivity during drought stress. Crop Sci. 2015;55:2012-33.
- [58]Robins JG, Bauchan GR, et al. Genetic mapping forage yield, plant height, and regrowth at multiple harvests in tetraploid alfalfa (Medicago sativa L.). Crop Sci. 2007;47:11-8.
- [59]Li X, Wei Y, et al. Association mapping of biomass yield and stem composition in a tetraploid alfalfa breeding population. Plant Genome. 2011;4.
- [60]Sakiroglu M, Sherman-Broyles S, et al. Patterns of linkage disequilibrium and association mapping in diploid alfalfa (M. sativa L.). TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2012;125:577-90.
- [61]Yu LX, Zheng P, et al. The impact of genotyping-by-sequencing pipelines on SNP discovery and identification of markers associated with verticillium wilt resistance in autotetraploid alfalfa (Medicago sativa L.). Front Plant Sci. 2017;8:89.
- [62]Musial JM, Mackie JM, et al. Identification of QTL for resistance and susceptibility to Stagonospora meliloti in autotetraploid lucerne. Theor. Appl. Genet.. 2007;114:1427-35.
- [63]Mackie JM, Musial JM, et al. Identification of QTL for reaction to three races of Colletotrichum trifolii and further analysis of inheritance of resistance in autotetraploid lucerne. Theor Appl Genet.. 2007;114:1417-26.
- [64]Brouwer DJ, Duke SH, et al. Mapping genetic factors associated with winter hardiness, fall growth, and freezing injury in autotetraploid alfalfa. Crop Sci. 2000;40:1387-96.
- [65]Narasimhamoorthy B, Bouton JH, et al. Quantitative trait loci and candidate gene mapping of aluminum tolerance in diploid alfalfa. Theor Appl Genet.. 2007;114:901-13.
- [66]McCord P, Gordon V, et al. Detection of QTL for forage yield, lodging resistance and spring vigor traits in alfalfa (Medicago sativa L.). Euphytica. 2014;200:269-79.
- [67]Wang X, Gao Y, et al. (E)-β-farnesene synthase gene affects aphid behavior in transgenic Medicago sativa. Pest management science. 2019;75:622-31.
- [68]Strizhov N, Keller M, et al. A synthetic cryIC gene, encoding a Bacillus thuringiensis delta-endotoxin, confers Spodoptera resistance in alfalfa and tobacco. Proc Natl Acad Sci U S A. 1996;93:15012-7.
- [69]Kang P, Bao AK, et al. Assessment of Stress Tolerance, Productivity, and Forage Quality in T(1) Transgenic Alfalfa Co-overexpressing ZxNHX and ZxVP1-1 from Zygophyllum xanthoxylum. Front. Plant Sci. 2016;7:1598.
- [70]Suárez R, Calderón C, et al. Enhanced tolerance to multiple abiotic stresses in transgenic alfalfa accumulating trehalose. Crop Sci. 2009;49:1791-9.
- [71]McKersie BD, Bowley SR, et al. Winter survival of transgenic alfalfa overexpressing superoxide dismutase1. Plant Physiol. 1999;119:839-48.
- [72]Bao A-K, Wang S-M, et al. Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci. 2009;176:232-40.
- [73]Reddy MS, Chen F, et al. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci U S A. 2005;102:16573-8.
- [74]Barone P, Rosellini D, et al. Bacterial citrate synthase expression and soil aluminum tolerance in transgenic alfalfa. Plant Cell Rep. 2008;27:893-901.
- [75]D'Halluin K, Botterman J, et al. Engineering of herbicide-resistant alfalfa and evaluation under field conditions. Crop Sci. 1990;30:866-871.
- [76]Wang Z, Ke Q, et al. Transgenic alfalfa plants expressing the sweetpotato Orange gene exhibit enhanced abiotic stress tolerance. PLoS One. 2015;10:e0126050.