Molecular characterization and phylogenetic analysis of two minnows, Puntius sarana and Barbodes gonionotus.
Abstract
Two minnows, indigenous olive barb, Puntius sarana and exotic silver barb, Barbodes gonionotus are important fish species in Bangladesh. Therefore, it is essential to identify diversed population of these fish species for selective breeding programme. Sixty olive barb fish were collected from three different natural stocks (Mymensingh, Madaripur and Sylhet) and 20 silver barbs from hatchery stock of Jashore in Bangladesh. Out of 40 decamer primers tested, 5 primers were selected for the Polymerase Chain Reaction (PCR) based RAPD (Randomly Amplified Polymorphic DNA) analysis. Upon agarose gel electrophoresis, RAPD bands were scored as separately on the basis of their presence or absence for each sample and primer. A total of 43 polymorphic bands and highest proportion of polymorphic bands (62.79%) were found in the Madaripur populations. The gene diversity (0.2132±0.2067) and Shannon’s Information index (0.3161±0.2950) within populations were highest in Jashore stock. Among olive barb stocks, these values were higher in Sylhet stock. Besides, four populations segregated in two main clusters based on the Nei’s genetic distance. Indicating the segregation of two different species of minnows, the silver barb stock made one separate cluster while other three stocks of olive barb remained in another cluster. The present study exposed a distinct pattern of genetic variation and phylogenetic relatedness that would be helpful in selecting broodfish for genetic improvement as well as in conservation of these fish species.
INTRODUCTION
Olive barb, Puntius sarana (Hamilton, 1822) and silver barb, Barbodes gonionotus (Bleeker, 1850) are two important minnows which are tropical and small freshwater fish belonging to the family Cyprinidae. The olive barb is a widely distributed cyprinid in the inland waters of South-East Asia [1] and is used both as food fish and ornamental fish. This species is omnivorous and feeds on aquatic weeds, algae, protozoan, mud and sand [2]. The natural abundance of olive barb has been reduced due to habitat fragmentation, injudicious usage of fertilizers in agricultural fields and their effluents mixing to water bodies and over-exploitation of water resources in Bangladesh and it is considered under vulnerable group in India [3; 4]. On the other hand, silver barb (Thai sarpunti), B. gonionotus, a herbivorous [5] and exotic fish introduced from Thailand to Bangladesh in 1977 that has been popular for its rapid growth, bright silvery outlook and taste [6]. Among all the exotic fish species it becomes one of the suitable species for aquaculture owing to its high yield potential and market demand [6].
Genetic conservation and broodstock selection of a fish species may be helpful for good practice by breeders and hatchery owners. In these aspects, genetic methods have great potential to distinguish populations of fish species that cannot be identified by morphological and meristic characters [7]. Data on genetic variation reflects the genetic condition of a species that can be used in designing proper selective breeding program for genetic improvement. DNA markers provide valuable and realistic genetic data that would be useful for investigation and monitoring of genetic conditions both in natural populations and in captive stocks. There are several types of DNA markers available among which PCR (Polymerase Chain Reaction) based RAPD (Randomly Amplified Polymorphic DNA) technique is very simple and quick to perform. The most important one that makes it simple from others technique, the genome specific sequence of the target organism is not required to design RAPD primers and only single primer is sufficient to amplify analyzable DNA fragments [8; 9]. RAPD technique was successfully applied for phylogenetic studies [10; 11], identification of subspecies [12] and gene mapping studies of fish species [13]. The objective of this research is to characterize different populations of P. sarana and one stock of B. gonionotus at molecular level and to indicate phylogenetic relatedness among the stocks of minnows in Bangladesh.
MATERIALS AND METHODS
Collection of fish sample and isolation of genomic DNA
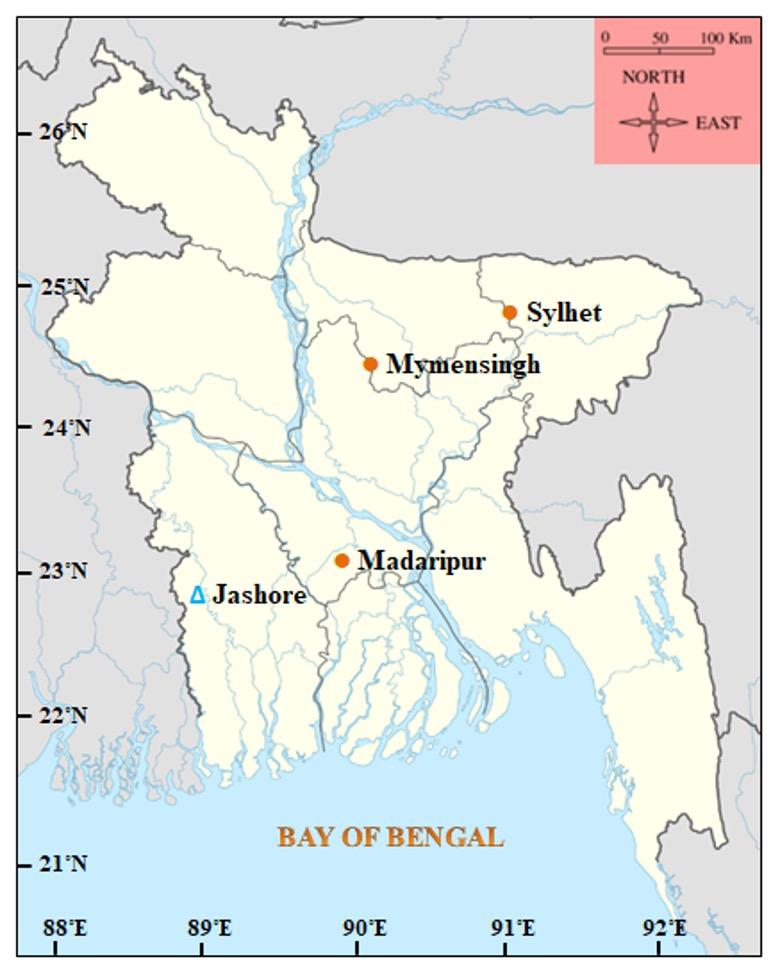
Indigenous olive barb fish samples (n= 60, 20 from each location) were collected from three sources namely Mymensingh, Madaripur and, Sylhet (Hakaluki Haor) in Bangladesh. The silver barb fish samples (n=20) were collected from a hatchery stock situated in Jashore district of Bangladesh. The locations of fish sampling are shown in the map of Bangladesh (Figure 1). The samples were collected during July 2015 to March 2016. From individual fish, a small piece of caudal fin was cut with scissors and preserved in 95% alcohol. Genomic DNA was isolated from the caudal fin according to the method described by Islam and Alam [14] and stored at -20 ºC.
Primer selection and PCR amplification
Primarily 40 decamer primers of random sequence (Bioserve Biotechnologies India Pvt Ltd. ACGI Company) were screened with one sample from each stock to test the performance of RAPD primers and resolution of bands generated. Finally five primers exhibiting highest quality banding patterns and sufficient variability were selected for analysis of all samples. DNA amplification was performed in a final volume of 25μl containing 12.5μl Master mix (Taq DNA Polymerase 2X-premix, GeneON), 2μl of template, 2μl of primer and 8.5μl of nuclease free water. The amplification was carried out in thermal cycler (Prime G Thermal Cycler, UK) programmed for initial heat denaturation in one step of 2 minutes at 94ºC. Subsequent 45 cycles of 1 minute at 94ºC, 30 seconds at 34ºC to 42ºC (OPA12: 38ºC, OPA17: 40ºC, OPB03: 34ºC, OPB07: 36ºC, OPB20: 42ºC) and 2 minutes at 72ºC; followed by one final step of primer extension at 72ºC for 7 minutes.
Agarose gel electrophoresis
PCR product (10μl) was subjected to electrophoresis in 1.5% agarose gel containing ethidium bromide on 1X TBE buffer at 100 V for 1 hour. Molecular marker (1 Kb Plus) was used alongside of the sample. Finally, DNA bands were observed on a GelDoc system and photographs were recorded.
RAPD data Analysis
The obtained bands were scored separately on the basis of their presence (1) or absence (0) for each sample and primers. For accuracy, the scoring was done by two persons independently. The total scores were then used to create a single data matrix to estimate the proportion of polymorphic loci, Nei’s gene diversity [15], gene flow and Nei’s genetic distance [16] and to construct an unweighted pair group method of arithmetic mean (UPGMA) dendrogram among stocks with 1,000 simulated samples using the POPGENE (Version 1.31) program [17]. Band sizes of the RAPD marker were estimated by using the software DNAfrag (Version 3.03) [18].
RESULTS
A total of 43 bands were scored and all of them are polymorphic (100% polymorphism) across all populations (Table 1). Number of bands ranged from 7 to 10 with an average 8.6 per primer. The highest (62.79%) polymorphic loci were found in Madaripur population and the lowest (51.16) in Mymensingh populations both of which are natural populations of olive barb (Table 2).The gene diversity was highest (0.2132±0.2067, Mean± SD) in Jashore followed by Sylhet (0.2021±0.2134), Madaripur (0.1850± 0.1883) and the Mymensingh (0.1597±0.1798) populations (Table 2). The Shannon’s Information index was also highest (0.3161±0.2950) in Jashore stock. In this research, the highest gene flow was found between Madaripur and Mymensingh populations and the lowest between Sylhet and Jashore populations (Table 3).
Table 1. Number and percentage of polymorphic loci of each RAPD marker tested in populations of olive barb P. sarana and silver barb, B. gonionotus.
Table 2. Number and percentage of polymorphic loci of each population of olive barb P. sarana and silver barb, B. gonionotus analyzed by five RAPD Primers.
Table 3. Gene flow between populations of olive barb, P. sarana and silver barb, B. gonionotus analyzed by five RAPD primers
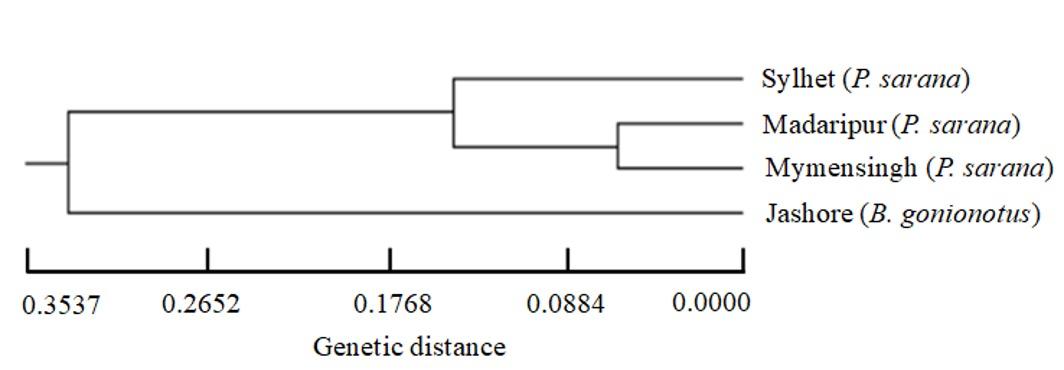
The genetic distance between Sylhet and Jashore populations was highest (0.3537) and was lowest (0.0559) between Madaripur and Mymensingh populations (Table 4). Both the values for genetic identity and gene flow between Madaripur and Mymensingh populations were highest. Based on the Nei’s genetic distance, the UPGMA dendrogram indicated the segregation of all four populations into two main clusters (Figure 2). The three natural and indigenous barb (Sylhet, Mymensingh and Madaripur) populations made one cluster and exotic silver barb (Jashore) remained in another cluster. The Madaripur and Mymensingh populations made one sub cluster and Sylhet population belonged to another sub cluster.
Table 4. Nei’s (1972) genetic identity (above diagonal) and genetic distance (below diagonal) of olive barb, P. sarana and silver barb, B. gonionotus analyzed by five RAPD primers.
DISCUSSION
The P. sarana is a small indigenous species (SIS) in Bangladesh. Once the fish was available in natural shallow water bodies including rivers, beels, haors in Bangladesh but with increase of pollution, overharvesting, introduction of alien species makes it restricted in some specific region the country. Information on the genetic structure of fish species is useful for optimizing identification of stocks, stock enhancement, breeding programs, management for sustainable yield and preservation of genetic diversity [19; 20; 21; 22]. Besides, the exotic Thai silver barb became a vital candidate for aquaculture in Bangladesh due to its high and fast growth rate over the indigenous olive barb. RAPD fingerprinting offers a rapid and efficient method for generating a new series of DNA markers in fishes [23].
DNA polymorphisms have been extensively employed as a means of assessing genetic diversity in aquatic organisms. In the present study polymorphism was 100% across all the primers. The highest proportion (62.79%) of polymorphic band was found Madaripur natural population. Akter et al. [24] found 43.34% polymorphic loci in a hatchery stock of olive barb where the brood fish were collected from natural sources (a haor of Sunamgonj district) and the samples were first generations. Kabir et al. [25] reported 53.84% polymorphism by RAPD marker analysis in three populations of P. sarana which were collected from natural sources where the primers were different from our study. The mean number of polymorphic loci was (26.6) found by Parvez et al. [26] in the study of stock genetic variation of natural stock of critically endangered P. sarana through allozyme electrophoresis. The higher polymorphic loci (42.31%) was reported in natural population of Labeo kalbasu by Mostafa et al. [27] used by RAPD marker. In this study, we also found the higher polymorphic loci in natural stock. The gene diversity was found to be higher (0.2132±0.2067) in the Jashore population which is the indication of higher genetic variation of this stock and it was a hatchery and silver barb stock. The higher genetic variation parameters can be used in the selective breeding programs. The higher gene diversity (0.3703±0.114, mean±SD) was found in natural population (Mohongonj haor) by Sultana et al. [28] in Heteropneustes fossilis fish. Gopalakrishnan et al. [29] found the mean gene diversity of 0.1848 from the study of genetic differentiation of Malabar carp Labeodus sumieri revealed by five RAPD primers. The highest genetic identity (0.9456) between Madaripur and Mymensingh populations indicates the same gene shared by these two populations though they are geographically isolated. The gene flow (3.8236) was also highest between these two populations. However, Jashore population maintained minimal gene flow with the other three populations. Since, samples collected from Jashore were of a captive stock of silver barb and are of a unique species, it is usual that there might have lower gene flow with the other three stocks of olive barb species.
Nei’s genetic distance was highest between Sylhet and Jashore populatios. In phylogenetic dendrogram, the Jashore population made one cluster and the remaining Sylhet, Madaripur, Mymensingh populations made another cluster which clearly indicated the segregation of two different species of minnows. Since Jashore population represents a captive stock of silver barb, it can be maintained in the hatchery for brood stock management in sustainable manners. Among the olive barb populations, Sylhet population showed better genetic diversity, because this fish population belongs to the large inland water bodies, Hakaluki haor in the eastern region of Bangladesh. As olive barb is critically endangered, our present study recommends conserving this population to protect from extinction of the species.
It can be concluded that remarkable level of genetic variable parameters like polymorphic loci, gene diversity were observed in the present study. However, these parameters can be taken into consideration for raising genetically superior broodstock in selective breeding program as well as for conservation management of these species of minnows family.
AUTHOR CONTRIBUTIONS
SS, MSH and MSI performed the experiment; SS, MS and JA conceived the study; SS and MNI analyzed the data; SS, MSH, MSI, MNI and JA wrote the paper.
CONFLICT OF INTEREST
The authors declare that no conflicts of interest exist.
References
- [1]Jena JK, Das PC, Das R, Mondal S. Performance of olive barb, Puntius sarana (Hamilton) in fingerling rearing with rohu, Labeo rohita (Hamilton) and mrigal, Cirrhinus mrigala (Hamilton). Aquacult 2007: 265(1): 305-308.
- [2]Mookerjee HK, Gupta SNS, Choudhury DNR. Food and its percentage composition of the common adult food fishes of Bengal. Sci Cult 1946; 47(12): 247.
- [3]Mijkherjee, M, Praharaj A, Das S. Conservation of endangered fish stocks through artificial propagation and larval rearing technique in West Bengal, India. Aquacul Asia 2002; 7(2): 8-11.
- [4]Chakraborty BK, Miah MI, Mirza M JA, Habib MAB. Rearing and nursing of local Sarpunti, Puntius sarana,(Hamilton) at different stocking densities. Pakistan J Biol Sci 2003; 6 (9): 797—800.
- [5]Haroon AKY, Pittman KA. Rice-fish culture: feeding, growth and yield of two size classes of Puntius gonionotus Bleeker and Oreochromis spp. in Bangladesh. Aqucult 1997; 154(3-4): 261-281.
- [6]Islam MS, Azim ME, Rahman MM, Mazid MA. Culture prospects of sharpunti (Puntius gonionotus Bleeker) and common carp (Cyprinus carpio Linnaeus) in backyard ditches at farmer level. Prog Agricult 1998; 8(1-2): 191-194.
- [7]Cadrin SX, Friedland KD, Waldman J. (Ed.). Stock identification methods: Applications in fishery science. Elsevier Academic Press, Burlington, MA 01803, USA, 2005; 719 pp.
- [8]Welsh J McClelland M. Fingerprinting genome using PCR with arbitrary primers. Nucleic Acids Res 1990; 18: 7213–7218.
- [9]Williams JGK, Kublik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 1990; 18: 6531–6535.
- [10]Bardakci F, Skibinski DOF. Application of the RAPD technique in tilapia fish: species and subspecies identification. Heredity, 1994; 73: 117–123.
- [11]Sultmann H, Mayer WE, Figueroa F, Tichy H, Klein J. Phylogenetic analysis of cichlid fishes using nuclear DNA markers. Mol Biol Evol 1995; 12: 1033–1047.
- [12]Partis L, Wells RJ. Identification of fish species using random amplified polymorphic DNA (RAPD). Mol Cell Probes 1996; 10: 435–441.
- [13]Postlethwait JH, Johnson SL, Midson CN, Talbot WS, Gates M, Ballinger EW, Africa D, Andrews R, Carl T, Eisen JS. A genetic linkage map for the zebrafish. Science 1994; 264: 699–703.
- [14]Islam MS, Alam MS. Randomly amplified polymorphic DNA analysis of four different populations of the Indian major carp, Labeo rohita (Hamilton). J Applied Ichthyol 2004; 20(5): 407-412.
- [15]Nei M. Analysis of gene diversity in sub divided populations. Proceedings of the Nationall Academy of Sciences USA 1973.
- [16]Nei M. Genetic distance between populations. American Naturalist 1972; 106: 283-292.
- [17]Yeh FC, Yang RC, Boyl T. POPGENE version 1.31.Microsoft window-based freeware for population enetic analysis.University of Alberta, Edmonton, Canada 1999.
- [18]Nash JHE. DNAfrag, Version 3.03. Institute for Biological Sciences, National Research Council of Canada, Ottawa, Ontario, Canada 1991.
- [19]Dinesh KR, Lim TM, Chua KL, Chan WK, Phang VPE. RAPD analysis: an efficient method of DNA fingerprinting in fishes. Zool Sci 1993; 10(5): 849-854.
- [20]Garcia DK, Benzie JAH. RAPD markers of potential use in penaeid prawn (Penaeus monodon) breeding programs. Aquacult 1995; 130(2-3): 137-144.
- [21]Tassanakajon A, Pongsomboon S, Rimphanitchayakit V, Jarayabhand P, Boonsaeng V. Random amplified polymorphic DNA (RAPD) markers for determination of genetic variation in wild populations of the black tiger prawn (Penaeus monodon) in Thailand. Mol Mar Biol Biotechnol 1997; 6(2): 110-115.
- [22]Tassanakajon A, Pongsomboon S, Jarayabhand P, Klinbunga S, Boonsaeng V. Genetic structure in wild populations of black tiger shrimp (Penaeus monodon) using randomly amplified polymorphic DNA analysis. J Mar Biotechnol 1998; 6: 249-254.
- [23]Foo C L, Dinesh KR, Lim TM, Chan WK, Phang VPE. Inheritance of RAPD markers in the guppy fish, Poecilia reticulata. Zool. Sci 1995; 12(5): 535-541.
- [24]Akter S, Sultana S, Khan MSR, Nahiduzzaman M, Hossain MAR, Alam MS. Genetic Characterization of Critically Endangered Puntius sarana (Hamilton) and the Exotic Barbonymus gonionotus (Bleeker)(Cyprinidae: Cypriniformes) by DNA Fingerprinting. Int J BioSci Agricult Technol 2010; 2(3): 21-27.
- [25]Kabir M, Habib M, Hossain A, Mandal SC. Genetic diversity of olive barb (Systomus sarana, Hamilton, 1822) from different locations of Bangladesh. Croatian J Fish 2015; 73(1): 6-12.
- [26]Parvez I, Alam MA, Amin AR, Islam MR, Khan MMR. Genetic variation and differentiation of wild stocks of critically endangered Puntius sarana (Hamilton) and their F1 crossbreed through allozyme electrophoresis. Int J Biosci 2015; 7(5): 47-57.
- [27]Mostafa MG, Ahmed ASI, Mustafa MG, Rabbane MG, Islam MN, Rafiquzzaman SM. Genetic diversity of wild and farmed Kalibaus (Labeo calbasu, Hamilton, 1822) by RAPD analysis of the genomic DNA. Croatian J Fish 2009; 67(2): 41-52.
- [28]Sultana S, Akter S, Hossain MAR, Alam MS. DNA fingerprinting of the Asian stinging catfish (Heteropneustes fossilis, Bloch) by Random Amplified Polymorphic DNA markers. Int J Biotechnol Appl 2010; 2(2): 5-10.
- [29]Gopalakrishnan A, Musammilu KK, Basheer, VS, John L, Padmakumar KG, Lal KK, Ponniah AG. Low genetic differentiation in the populations of the Malabar carp Labeo dussumieri as revealed by allozymes, microsatellites and RAPD. Asian Fish Sci 2009; 22(2): 359-391.