Screening of antagonistic potential bacteria from rhizosphere soil against phytopathogenic fungi related to selected vegetable crops
Abstract
Fungal phytopathogens cause serious losses of crop production worldwide, which causes serious losses of crop production. Bacteria play a role as the biocontrol agents for plant disease control. For this reason, the present study was conducted to determine the antagonistic potential of rhizosphere soil bacteria against selected phytopathogenic fungi. The screenings of potential antagonist isolated bacteria were applied by the dual culture technique with fungi Fusarium oxysporum and Colletotrichum melongenae. Molecular characterization was performed through 16S rDNA sequencing analysis. Sixteen (16) out of fifty (50) isolated bacteria showed different degrees of antagonism (25-67%) against both fungi F. oxysporum and C. melongenae. Among them, four (4) isolated bacteria (isolates A4, C1, C2, and E2) exhibited strong antagonism (more than 50% mycelial growth inhibition) against both fungi. The 16S rDNA sequences of the isolated bacteria A4, C1, C2 and E2 were 99-100% similar to Providencia sp. TT14, Bacillus subtilis 168, Bacillus subtilis RKP-2, and Bacillus amyloliquefaciens IBSDG-11, respectively. Based on the capability for the control of mycelial growth against both fungi, B. subtilis IUBTC2 was selected for optimization of growth characteristics and identification the bioactive metabolites which can enhance plant growth and disease control capacity of plants against the phytopathogenic fungi.
Keywords
INTRODUCTION
Major crop production losses are caused worldwide by pathogenic microorganisms which is comparable to approximately $220 billion lost per year [1]. Worldwide, the crop production damage was also caused by different fungal phytopathogenic species as Fusarium, Rhizoctonia or Colletotrichum such [2-5]. Fusarium wilt of cucumber diseases was caused by F. oxysporum, which is a very common disease in cucumber worldwide and causes serious losses every year [6-8]. Until now, disease control mechanism mostly depends on the practice of artificial fungicides. This approach is very challenging, because fungal stains may become resistant to fungicides due to the accumulation of these artificial fungicide’s in the ecosystem [9, 3]. Artificial fungicides are expensive. So, these are not sufficient for the disease control strategies [10].
In addition, public anxiety related to chemical pesticides has raised key attention to pursue additional control approaches [11]. Interactions are prevalent in nature between plant pathogens and antagonistic microorganisms. Similarly, plant pathogenic fungi and antagonistic microorganism interactions can employ to control or reduce diseases as biocontrol/biological agents which can suppress the plant pathogens [12]. It is one of the paths to reduce the usage of artificial fungicides in agriculture sector, where plant protection from diseases can be done by using microorganisms [13]. The biocontrol or biological control mechanisms are the secretion of both small molecules and enzymes, which are extremely active against pathogens [14, 1, 3].
Antagonism is the phenomenon in which one microorganism inhibits the other interacting partner to ensure its own survival. The stalling products of microbes are preferably called antibiotic substances or metabolites. It was reported that antagonistic bacteria act as a biocontrol agent due to their ability of the different modes of action [12]. The antagonistic effect of microorganism is the result of interaction among the microbial populations. Several bacterial species have been tested as biocontrol/biological agents [15]. The bacterial antagonisms mechanisms are involved in competition for space or nutrients and antibiosis for enrichment of plant and root growth, and for introduction of plant inactivation and/or resistance of the pathogen’s enzymes [16].
Antibiosis is the utmost significant machinery for the plant disease control. Cucumber (Cucumis sativus L.) is one of the key significant economical crops, which is grown either in the open field or greenhouses [17, 18]. Fusarium wilt of cucumber, caused by F. oxysporum f. sp. cucumerinum, is a very common disease of cucumber and oil palm in the world and causes serious losses every year [19, 6]. Eggplant (Solanum melongena) is a tropical, herbaceous, perennial plant, in the family Solanaceae, which is grown for its edible fruit [20]. The marketable eggplant production is negotiated due to affect by the insect which are generally recognized such as eggplant shoot and fruit borer (Leucinodes orbonalis) as well as various phytopathogenic fungi namely, Fusarium spp., C. melongenae, Pythium spp., Leveillu lataurica, Rhizoctonia spp., and Verticillium spp. [17]. Until now, disease control mainly deepens on the usage of synthetic fungicides. For this reason, the objective of this study was to screen the antagonistic potential of bacteria from rhizosphere soil against phytopathogenic fungi in selected vegetable crops in Bangladesh. At first, we isolated bacteria from rhizosphere soil at different sources and screened the antagonistic potential bacteria against phytopathogenic fungi. We also further investigated as morphological, biochemical and molecular characterization to determine the biocontrol/biological agents for the phytopathogenic fungus control.
MATERIALS AND METHODS
Fungal pathogens
Two fungal pathogens, namely, F. oxysporum cucumerinum and C. melongenae were used in this study, which were obtained from the Laboratory of Microbiology, Islamic University, Kushtia-7003, Bangladesh.
Isolation of soil bacteria
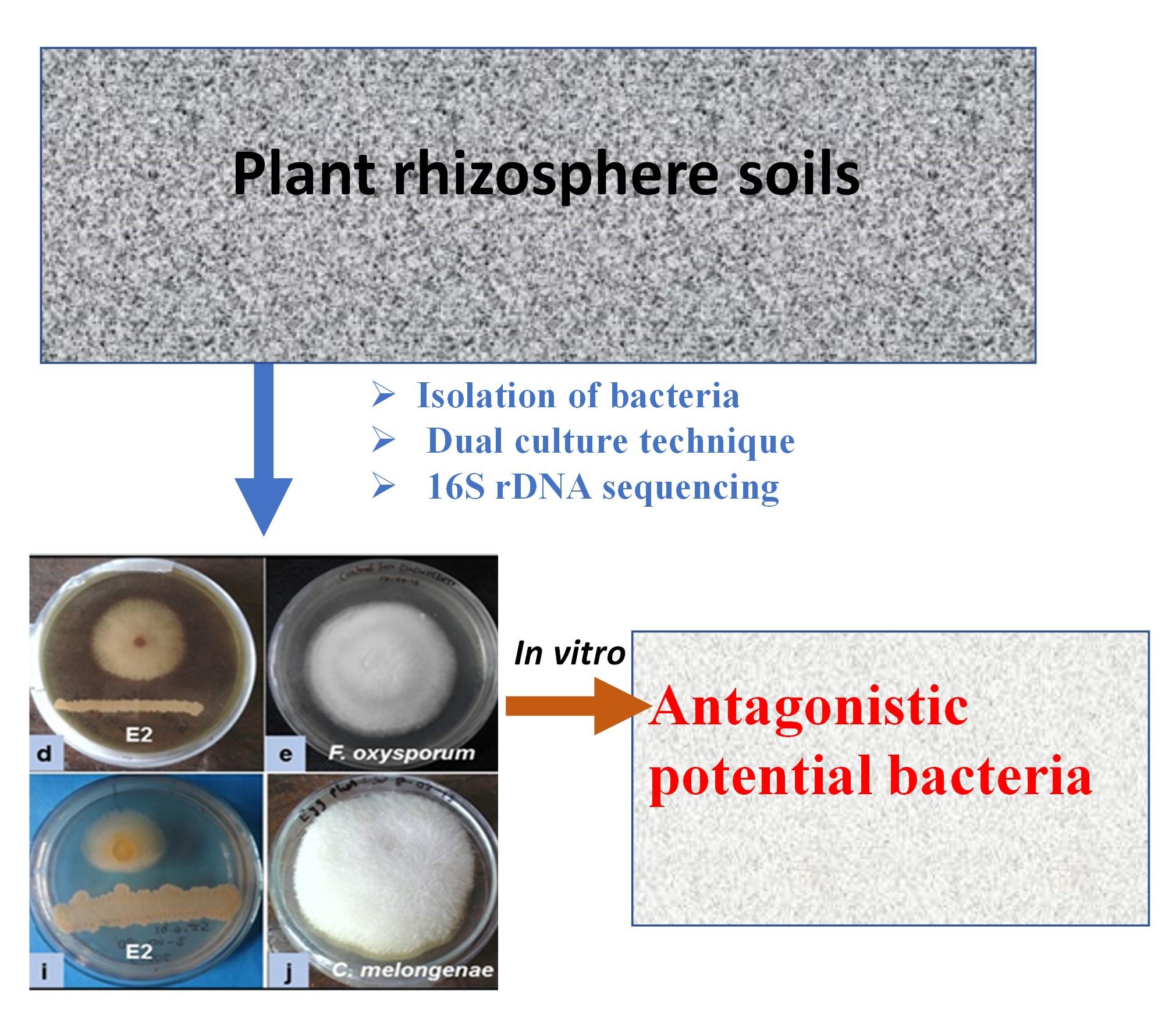
Rhizosphere soil was collected from 5 different agricultural and non-agricultural field crops of Shantidanga, Kushtia. The soil samples were brought to the laboratory in sterile polyethylene bags and stored at 4ºC for the isolation of microorganisms. Soil bacteria were isolated by serial dilution method from soil samples. In brief, it was suspended 5 g of soil in sterile distilled water and shaking at 120 rpm for 10 min in a rotary shaker. Then it was diluted with distilled water (1:9 ratio) up to107 fold. 100 μl aliquots from 104 to 107 dilutions was spread in the tryptone soya agar (TSA) media and mildly spread by a sterile glass spreader. Then it was incubated at 35ºC for 3 days, after it was sub-cultured onto the same medium plates based on morphological distinct colony and isolated single colonies from these plates.
Screening of antagonistic activities
Antagonistic activities were screened from the isolated bacterial strains through In vitro. Mycelial growth inhibition of F. oxysporum and C. melongenae was achieved through the dual culture method in potato dextrose agar (PDA) media. PDA media was organized and transferred in petri dishes (20 ml). Agar plug (5 mm) of vigorously rising culture of each fungus was positioned into the center of individual plate. Individual isolate was streaked 3 cm away and 1 cm away from the “edge of the” petri dish. Plates without bacterial inoculation was used a control. Then the plate was enclosed with parafilm and incubated at a temperature (27ºC) for 5 days till the fungal mycelia touched the control plate edge. All tests were done in triplicate for each isolate. Then circular mycelial development, inhibition percentage (%) was measured by the following formula [21]:
Radial inhibition (RI%)=(A1-A2)/A1x100
Where, A1 = radial growth of fungal mycelia without bacterial isolate, A2 = radial growth of fungal mycelia with bacterial isolate.
Morphological characterization and biochemical test of isolated bacteria
The morphological characterizations and physiological and biochemical tests were done on the selected isolates. In brief, morphological characterizations such as size, shape, motility and spore forming were observed. The biochemical tests such as Gram staining, catalase test, KOH test, lactose fermentation, sulfur reduction, and urease test of the isolated bacteria were done as described earlier [22, 23]. Determination of degree of growth of isolated bacteria, different types of media were used such as tryptone soya agar (TSA), lactose broth agar (LBA), yeast extract agar (YEA), potato dextrose agar (PDA), and King’s medium B agar (KBA).
Molecular characterization of the isolated bacteria
Genomic DNA was extracted from the selected isolated bacteria according to the protocol designated by He [24] (phenol: chloroform: iso-amyl alcohol procedure). The sequence of 16S rDNA was amplified through polymerase chain reaction (PCR). Thermal cycles and other conditions were applied according to Rahman et al [25, 26]. The universal primer set of 63F (5-CAGGCCTAACACATGCAAGTC-3) [25, 26, 27] and 1389R (5-ACGGGCGGTGTGTACAAG-3) [25, 26, 28] was used for the amplification of 16S rDNA. Amplified PCR products were purified by purification kits (TaKaRa, Hot-Start Version). Then the purified PCR products were sequenced by DNA sequencer (Invent Technology, Dhaka, Bangladesh). The obtained sequences were analyzed through Chromas software. The similarity was observed with known sequences present in the NCBI GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) through the BLAST program. Multiple sequence alignment (MSA) of 16S rDNA sequences of the isolated bacteria was performed by using the Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The aligned sequences were then exposed to a Neighbor-Joining phylogenetic tree constructed by Clustal Omega. Then 16S r DNA sequences were deposited into the NCBI data bank. The sequences were submitted in the NCBI gene bank and accession numbers as MG237885 to MG237888 for Providencia sp. IUBTA4, B. subtilis IUBTC1, B. subtilis IUBTC2, and B. amyloliquefaciens IUBTE2, respectively.
RESULTS
Screening of antagonistic activities from isolated bacteria through an in vitro assay
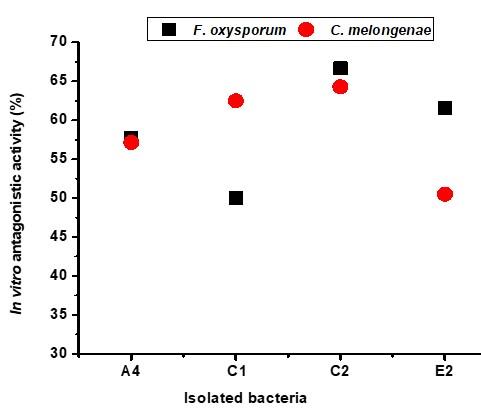
Screening of antagonistic activity of each isolated bacterium was accomplished through an in vitro assay as shown in Figure 1. Fifty (50) bacterial isolates were obtained from five (5) different soil samples. Some isolates were antagonists against only one fungus. Among them, sixteen (16) isolates showed different degrees of antagonism (25-67%) against both fungi, F. oxysporum and C. melongenae. Four (4) isolates A2 (58% and 57%), C1 (50% and 63%), C2 (67% and 64%), and E2 (62% and 50%) showed a higher efficacy in inhibiting the radial growth fungal mycelia. The strong antagonism (more than 50% mycelial growth inhibition) was exhibited by the isolates A4, C1, C2, and E2 against both fungi (Figure 2).
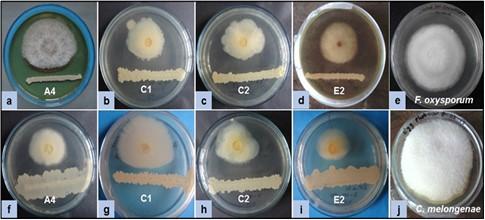
Morphological characterization of isolated bacteria
Morphological characterizations of isolated bacteria are shown in Table 1. Morphological studies showed that the isolated bacteria A4 was straight rods, non-spore former, and motile. The isolated bacteria C1, C2, and E2 were rods shape, spore former, and motile. The degree of growth characteristics of the isolated bacteria A4, C1, C2 and E2 was observed using different types of media as shown in Table 2 and the growth patterns were recorded as shown Figure 3. Four (4) isolated bacteria A4, C1, C2 and E2 showed the excellence growth and good growth on TSA and KBA media, respectively as compared with other media. The growth patterns revealed that there was a distinct difference among the isolated bacteria.
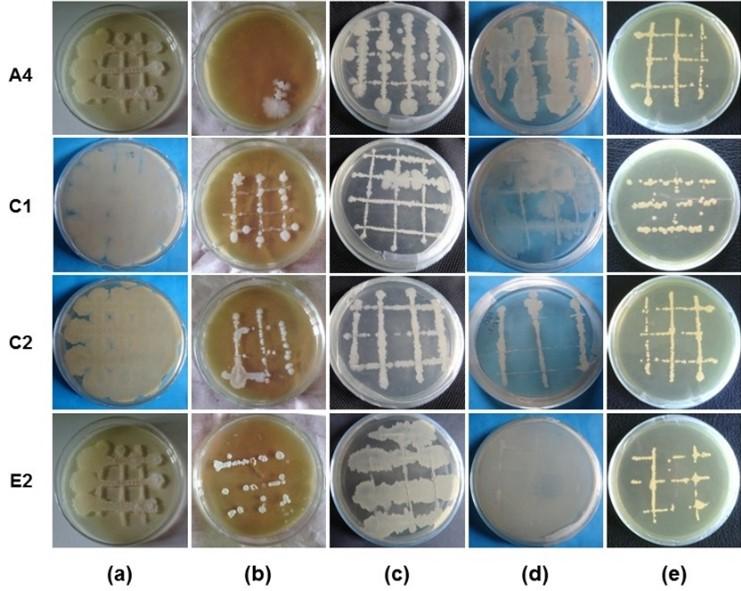
Table 1. Morphological characteristics and the biochemical properties of the selected isolated bacteria.
Table 2. Degree of growth of the selected isolated bacteria in different culture media.
Biochemical test of isolated bacteria
The results of biochemical test of isolated bacteria were shown in Table 2. The isolated bacteria C1, C2, and E2 were positive for the Gram staining and KOH test, respectively. Isolated bacteria A4 was negative against gram reaction and KOH test, respectively. Isolated bacteria A4, C1, C2, and E2 were positive in the catalase test and urease test. On the other hand, all of the four isolated bacteria showed negative for lactose fermentation and sulphur reduction (Table 2).
Molecular identification of selected bacterial isolated
Molecular identification was applied by 16S rDNA sequence analysis as shown in Table 3. The 16S rDNA sequences were received and analyzed in the Chromas software (version 2.6.4) showed good quality of the sequences. The similarity of the sequence of the isolates was exposed (100-99% similarity) through BLAST exploration and there was a close relationship presented between the isolated strains and known sequence of the gene bank NCBI. Isolated bacteria A4, C1, C2 and E2 were most closely related to Providencia sp. BAB-5310, Bacillus sp. E157, B. subtilis subsp. inaquosorum strain PF9 and B. amyloliquefaciens strain BG2. The sequences were submitted in the NCBI Gene Bank and accession numbers A4 (MG237885), C1 (MG237886), C2 (MG237887) and E2 (MG237888) were received for the isolated bacteria. Finally, the four isolates were identified as Providencia sp. IUBTA4, B. subtilis IUBTC1, B. subtilis IUBTC2 and B. amyloliquefaciens IUBTE2. Neighbor-joining phylogenetic trees were constructed of isolated bacteria as shown in Figure 4. Phylogenetic analysis indicated that isolated bacteria were represented in a distinct class, demonstrating that they are newly isolated bacteria as compared to NCBI Data bank.
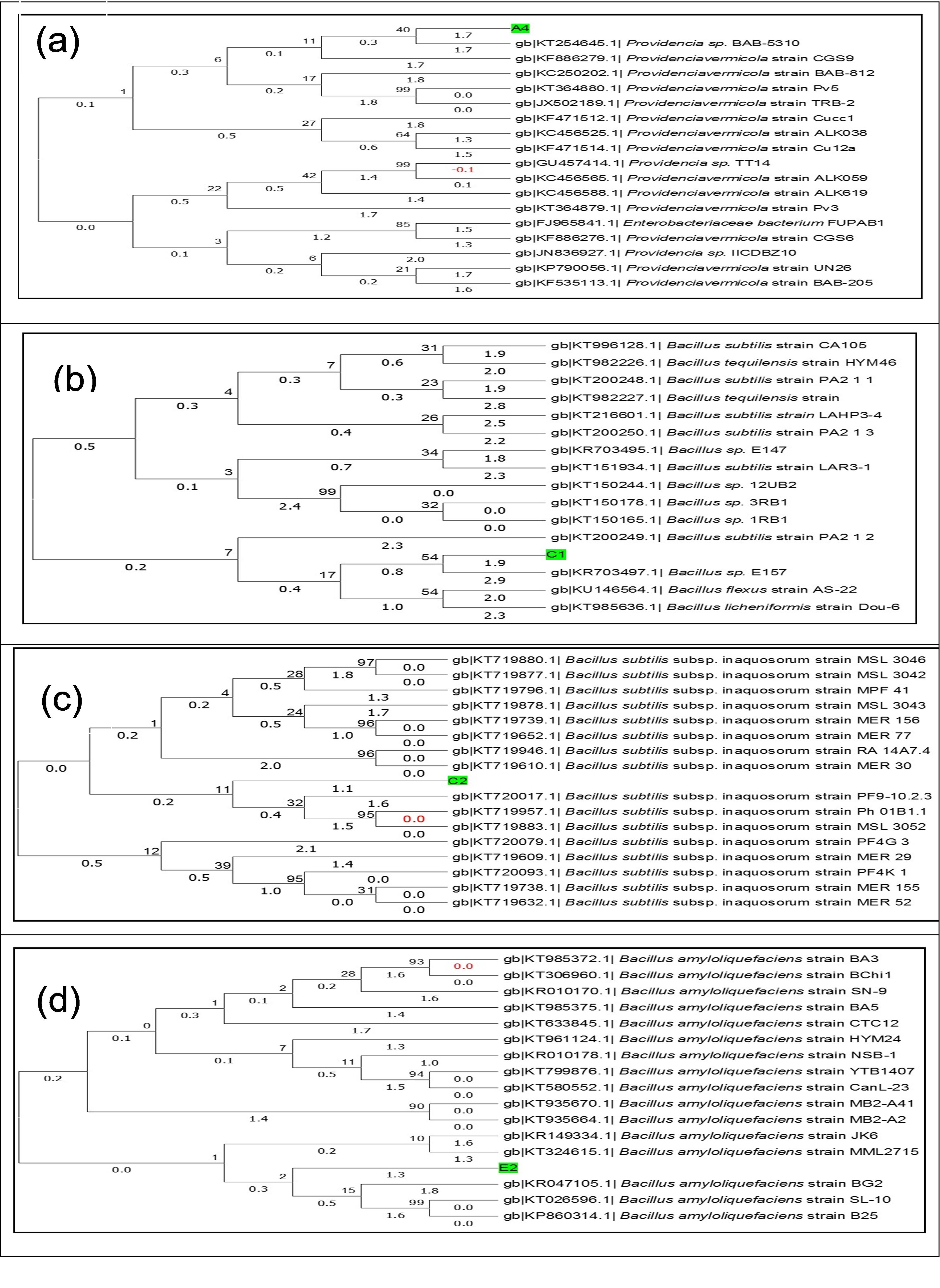
Table 3. Closely related species of the four selected isolated bacteria based on the similarity of the partial 16S rDNA sequences in BLASTN.
DISCUSSION
The use of valuable microorganisms considers the greatest auspicious approaches for the harmless crop management performs [29]. In this respect, we tried to isolate and identify antagonistic bacteria with robust antifungal activities against F. oxysporum and C. melongenae. In our investigation, we found that among fifty (50) isolated bacteria, sixteen (16) isolated bacteria showed different degrees of antagonism (25-67%) against both fungi in dual culture. Four isolates with an inhibition rate above 50% were selected for further investigations (Figure 2). In vitro dual culture examination was widely applied to the preliminary screening of biocontrol/biological agents [21]. Effects of antagonistic are frequently established by the development of inhibition zones between fungal and isolated bacteria [21] or by calculating the radial mycelial growth inhibition in percentage towards the isolated bacteria [30]. The selected isolates were identified based on morphological, biochemical and molecular techniques.
Molecular characterization was established and approved almost 2 decades years before to identify isolated bacteria species. This identification is commonly based on some sole parts of their subunit of 30S ribosomal RNA, which is called 16S rDNA [31]. So, 16S rDNA- sequence based techniques were extensively applied to characterize the bacterial community structure in soils [32, 25, 26, 33]. The 16S rDNA sequences of isolateA4, C1, C2, and E2 revealed that they were closely matched with Providencia sp, B. subtilis 168, B. subtilis RKP-2, and B. amyloliquefaciens, respectively (Table 3).
The B. subtilis are Gram positive, rod shaped, motile soil bacteria. Both B. subtilis strains showed the antagonistic activities in the dual culture. B. subtilis IUBTC1 showed the inhibition rates 50% and 63% against fungi F. oxysporum and C. melongenae respectively, whereas B. subtilis IUBTC2 showed the inhibition rates 67% and 64% against both fungi respectively due to produce more enzyme substances (Figure 2). Similarly, Burhan et al. [34] demonstrated that different species of Bacillus, most notably, B. subtilis produced approximately 60% of commercially available enzymes. In previous studies, Bacillus spp. exhibited very broad spectra of action with an efficient antagonistic activity against F. oxysporum [35-37]. Several species of Bacillus have been recognized as plant-growth promoting bacteria (PGPB) and/or biocontrol agents (BCA) [37]. It was also reported that the genus Bacillus has become a dependable choice to discovery out novel and promising bacteria for the making of amylase and other extracellular enzymes [33]. According to the NCBI database, isolate E2 was identified as B. amyloliquefaciens IUBTE2. It exhibited inhibition rates 62% and 50% against F. oxysporum and C. melongenae, respectively (Figure 2). In previous, it was reported that the non-pathogenic bacilli, B. amyloliquifaciens showed an inhibition rate of 46% against F. oxysporum [38]. Providencia spp. are as one of the most common multi-drug resistant bacteria [39]. Similarly, Godebo et al [40] reported that 75% of Providencia isolates were multi-drug resistant. In this study, isolated bacteria A4 was identified as Providencia sp. IUBTA4 that showed 58% and 57% mycelial growth inhibition against F. oxysporum and C. melongenae, respectively, due to biocontrol potential elicited defense enzymes (Figure 2). Similarly, Rana et al. [41] reported that Providencia spp. act as PGPR with biocontrol potential elicited defense enzymes in wheat.
CONCLUSION
The main ecological tasks facing microbiologists and plant pathologists are to produce the environment-friendly substitutions instead of chemical pesticides for the fighting of crop infections. Sixteen (16) isolates showed different degrees of antagonism (25-67%) against both fungi F. oxysporum and C. melongenae. Four (4) isolated bacteria, Providencia sp. TT14, B. subtilis 168, B. subtilis RKP-2, and B. amyloliquefaciens IBSDG-11, respectively were exhibited strong antagonism (more than 50% mycelial growth inhibition). B. subtilis IUBTC2 showed the highest capability for the control of mycelial growth against phytopathogenic fungi. Finally, it may conclude that newly isolated bacteria have a potential disease control capacity of the plant against the pathogenic fungi which can be enhanced plant growths and seed emergences as well as act as biocontrol agents for better green environmental management (Figure 5).
ACKNOWLEDGMENTS
The authors grateful to Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia-7003, Bangladesh for assistance of research activities.
AUTHOR CONTRIBUTIONS
MKA, MMR, and MRI comprehended and planned the study; MKA, and AHMJ carried out the analysis; MKA, and MMR wrote the manuscript; MMR prepared the graphs and illustrations; AHMJ and MRI contributed to the critical revision of the manuscript; MMR and MRI supervised the whole work; and all authors approved the final manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Kandel SL, Firrincieli A, Joubert PM, Okubara PA, Leston ND, McGeorge KM, et. al, An in vitro study of biocontrol and plant growth promotion potential of Salicaceae endophytes. Frontiers in Microbiology. 2017; 8:386.
- [2]Imran QM, Yun B. Pathogen-induced Defense Strategies in Plants. Journal of Crop Science and Biotechnology.2020; 23:97–105,
- [3]Mehnaz S, Saleem RSZ, Yameen B, Pianet I, Schnakenburg G, Pietraszkiewicz H, Harald G. Lahorenoic acids A-C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. Journal of Natural Products 2013;76(2):135–141.
- [4]Winterab M, Samuelsa PL, Donga Y, Dill-Mackya R. Trichothecene production is detrimental to early root colonization by Fusarium culmorum and F. graminearum in fusarium crown and root rot of wheat. Plant Pathology. 2019; 68:185–19.
- [5]Patel S, Rajput K, Saraf M. Elicitation of plant defense enzymes against Fusarium oxysporum f. sp. lycopersici in tomato plant using a novel rhizobacteria Providencia rettgeri MSS2. Biocatalysis and Agricultural Biotechnology. 2017; 12:308313.
- [6]Kazan K, Gardiner DM. Transcriptomics of cereal-Fusarium graminearum interactions: what we have learned so far. Mol. Plant Pathology. 2018; 19(3):764–778.
- [7]Das MM, Haridas M, Sabu A. Biological control of black pepper and ginger pathogens, Fusarium oxysporum, Rhizoctonia solani and Phytophthora capsici, using Trichoderma spp. Biocatalysis and Agricultural Biotechnology. 2019; 17:177–183.
- [8]Palacio-Barrera AM, Areiza D, Zapata P, Atehortúa L, Correa C, Peñuela-Vásquez M. Induction of pigment production through media composition, abiotic and biotic factors in two filamentous fungi. Biotechnology Reports. 2019; 21:e00308.
- [9]Hassan MN, Afghan S, Hafeez FY. Biological control of red rot in sugarcane by native pyoluteorin-producing Pseudomonas putida strain NH-50 under field conditions and its potential modes of action. Pest Management Science. 2011; 67(9):1147–1154.
- [10]Kumar A, Verma H, Singh VK, Singh PP, Singh SK, Ansari WA, Pandey KD. Role of Pseudomonas sp. in sustainable agriculture and disease management. In V. S. Meena, P. K. Mishra, J. K. Bisht, & A. Pattanayak (Eds.), Agriculturally Important Microbes for Sustainable Agriculture Singapore: Springer Singapore (2017). doi.10.1007/978-981-10-5343-6_7
- [11]Nicolopoulou-Stamati P, Maipas S, Kotampasi C, Stamatis P, Hens L. Chemical pesticides and human health: the urgent need for a new concept in agriculture. Frontiers in Public Health. 2016; 4 :148
- [12]Köhl J, Kolnaar R, Ravensberg WJ. Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Frontiers in Plant Science. 2019; 10: 845.
- [13]van Lenteren JC, Bolckmans K, Köhl J, Ravensberg WJ, Urbaneja A. Biological control using invertebrates and microorganisms: plenty of new opportunities. BioControl. 2018; 63: 39–59.
- [14]Eilenberg J, Hajek A, Lomer C. Suggestions for unifying the terminology in biological control. BioControl. 2001; 46:387–400.
- [15]Moita C, Feio SS, Nunes L, Curto MJM, Roseiro JC. Optimization of physical factors on the production of active metabolites by Bacillus subtilis 355 against wood surface contaminant fungi. International Biodeterioration & Biodegradation. 2005; 55(4): 261–269.
- [16]Harman GE. Myths and Dogmas of biocontrol changes in perceptions derived from research on Trichoderma harzinum T-22. Plant Disease. 2000; 84(4):377–393.
- [17]Islam MA, Nain Z, Alam MK. et al. In vitro study of biocontrol potential of rhizospheric Pseudomonas aeruginosa against Fusarium oxysporum f. sp. cucumerinum. Egyptian Journal of Biological Pest Control. 2018; 28:90.
- [18]Saravanakumar K, Yu C, Dou K, Wang M, Li Y, Chen J. Synergistic effect of Trichoderma-derived antifungal metabolites and cell wall degrading enzymes on enhanced biocontrol of Fusarium oxysporum f. sp. cucumerinum. Biological Control. 2016; 94:37–46.
- [19]Adusei-Fosu K, Dickinson M, Yankey EN. AFLP as a fingerprinting tool for characterising isolates of Fusarium oxysporum f. sp. elaeidis causal organism for fusarium wilt disease of oil palm in Ghana. Journal of Plant Diseases and Protection. 2019; 126:575–584.
- [20]Parvaiz M, Hussain K, Rafique Y, Haider A, Nasreen S, Kousar S. In vitro Techniques for Propagation of Brinjal (Solanum melongena L.). World Applied Sciences Journal. 2013; 27(8) :1071–1078.
- [21]Ji SH, Gururani MA, Chun SC. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiological Research. 2014; 169(1):83–98.
- [22]Rahman MM, Rahman MM, Akhter S, Jamal MA, Pandeya DR, Haque MA, Alam MF, Rahman A. Control of coliform bacteria detected from diarrhea associated patients by extracts of Moringa oleifera. Nepal Medical College Journal. 2010; 12:12–19.
- [23]Chomvarin C, Chantarasuk Y, Mairiang P. et al. Sensitivity and specificity of an in-house rapid urease test for detecting Helicobacter pylori infection on gastric biopsy. The Southeast Asian Journal of Tropical Medicine and Public Health. 2006; 37(2):312–9.
- [24]He F. E. coli Genomic DNA Extraction. Bio-protocol. 2011; 1(14). http://www.bio-protocol.org/e97
- [25]Rahman MM, Basaglia M, Favaro L, Boz B, Gumiero B, Casella S. A wooded riparian strip set up for nitrogen removal can affect the water flux microbial composition. Italian Journal of Agronomy.2014; 9(1): 33–37.
- [26]Rahman MM, Basaglia M, Vendramin E, Boz B, Fontana F, Gumiero B, Casella S. Bacterial diversity of a wooded riparian strip soil specifically designed for enhancing denitrification process. Biology and Fertility of Soils. 2014; 50: 25–35.
- [27]Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Applied and Environmental Microbiology.1998; 64:795–799.
- [28]Osborn M, Moore ERB, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environmental Microbiology. 2000; 2(1): 39–50.
- [29]Lim SM, Yoon MY, Choi GJ, Choi YH. et al. Diffusible and Volatile Antifungal Compounds Produced by an Antagonistic Bacillus velezensis G341 against Various Phytopathogenic Fungi. Plant Pathology Journal. 2017; 33(5): 488–498.
- [30]Lee T, Park D, Kim K, Lim SM, Yu NH, Kim S, Kim HY. et al. Characterization of Bacillus amyloliquefaciens DA12 showing potent antifungal activity against mycotoxigenic Fusarium species. Plant Pathology Journal. 2017; 33:499–507.
- [31]Lynch JM, Benedetti A, Insam H. et al. Microbial diversity in soil: ecological theories, the contribution of molecular techniques and the impact of transgenic plants and transgenic microorganisms. Biology and Fertility of Soils. 2004; 40:363–385.
- [32]Rahman MM, Sultana T, Ali MY, Rahman A. Chemical composition and antibacterial activity of the essential oil and various extracts from Cassia sophera L. against Bacillus sp. from soil. Arabian Journal of Chemistry. 2017; 10(2):2132–2137.
- [33]Dash BK, Rahman MM, Sarker PK. Molecular Identification of a Newly Isolated Bacillus subtilis BI19 and optimization of production conditions for enhanced production of extracellular amylase. BioMed Research International. 2015; 2015:859805.
- [34]Burhan A, Nisa U, G ̈okhan C, Omer C, Ashabil A, Osman G. Enzymatic properties of a novel thermostable, ther-mophilic, alkaline and chelator resistant amylase from an alka-liphilic Bacillus sp. isolate ANT-6. Process Biochemistry. 2003; 38: 1397–1403.
- [35]Adesina MF, Lembke A, Costa R, Speksnijder A, Smalla K. Screening of bacterial activity towards Rhizoctonia solani and Fusarium oxysporum: Site-dependent composition and diversity revealed. Soil Biology and Biochemistry. 2007; 39(11):2818–2828.
- [36]Ntushelo K, Ledwaba LK, Rauwane ME, Adebo OA, Njobeh PB. The Mode of Action of Bacillus Species against Fusarium graminearum, tools for investigation, and future prospects. Toxins (Basel). 2019;11(10):606.
- [37]Khan N, Martínez-Hidalgo P, Ice TA, et al. Antifungal activity of Bacillus species against Fusarium and analysis of the potential mechanisms used in biocontrol. Frontiers in Microbiology. 2018; 9:2363.
- [38]Yuan J, Raza W, Shen Q, Huang Q. Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Applied and Environmental Microbiology. 2012;78(16):5942-5944.
- [39]Linhares I, Raposo T, Rodrigues A, Almeida A. Frequency and antimicrobial resistance patterns of bacteria implicated in community urinary tract infections: a ten-year surveillance study (2000-2009). BMC Infectious Diseases. 2013;13: 19
- [40]Godebo G. Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Annals of Clinical Microbiology and Antimicrobials. 2013; 12: 17.
- [41]Rana A, Saharan B, Kabi SR, Prasanna R, Sain L. Providencia, a PGPR with biocontrol potential elicits defense enzymes in wheat. Annals of Plant Protection Sciences. 2011; 19(1) :138–141.