Prevalence of single-nucleotide polymorphism (-308G>A) in the TNF-α promoter region correlates coronary heart disease among type-2 diabetic patients from the northern region of Bangladesh
Abstract
Tumor necrosis factor-alpha (TNF-α) is a major cytokine for inflammatory response in human body. This is also well linked with obesity and causing different pathophysiological problems in type 2 diabetic mellitus (T2DM) patients because of its pro-inflammatory over expression. However, seemingly harmless nucleotide changes in the promoter region often cause oscillation in expression, results in complications like dyslipidemia and atherosclerosis that ultimately exploit to coronary heart disease (CHD). Therefore, this study was designed to evaluate association between TNF-α (-308G>A) polymorphism at promoter region and CHD in T2DM patients from the northern region (Rajshahi) of Bangladesh. The total number of participants was 96 T2DM patients. TNF-α polymorphism were detected using high resolution melting (HRM) curve analysis. A total of 32 participants were suffering from CHD and 8 polymorphism (3 homozygous and 5 heterozygous) were detected among them. From Fisher’s Exact test, we found significant (P < 0.05) relationship between TNF-α (-308G>A) polymorphism and CHD in T2DM patients. According to Kendall’s Tau correlation matrix (r = 0.218), there is a good correlation between target polymorphism and CHD. Therefore, the overall results suggest that TNF-α promoter (-308G>A) polymorphism influences CHD in T2DM patients.
INTRODUCTION
Coronary heart disease (CHD) is one of the leading causes of death throughout the world. It is a chronic inflammatory disease [1] and atherosclerosis is the core clinical sign of CHD [2]. Our immune system plays the key role in the progression of atherosclerotic process from beginning to plaque rupture [1]. The pro-inflammatory TNF-α is well known to be concerned in the etiology of CHD and to be an independent predictor of cardiovascular disease (CVD) [3, 1]. Inflammatory cytokines seem to provoke the expression of controllers involved in the development of atherosclerosis complications [4]. Imbalanced discharge of pro-inflammatory cytokines including TNF-α is regulated by a number of genes which are thought to be involved in the pathogenesis of CHD [2]. An elevated serum level of TNF-α is an autonomous predictor of CVD [5].
TNF-α gene is sited at 6p21.3 location on the human chromosome and is arranged within class III region of major histocompatibility complex [6]. The p38 mitogen-activated protein kinase maintains provocation of this gene [7].
TNF-α is also involved in the pathogenesis of numerous autoimmune diseases including rheumatoid arthritis, septic shock and other inflammatory disorders including T2DM [8, 9]. It has a very strong role in lipid metabolism and has been associated with insulin resistance and it causes changes in lipid and glucose level involved in CVD risk [10, 11]. In severely obese and insulin resistant fa/fa rats, TNF-α mRNA expression is increased in adipose tissue but in deficiency of TNF-α expression causes peripheral insulin sensitivity in obese mice [12]. There is a very concrete relationship between levels of TNF-α in adipose tissue and the scope of hyperinsulinaemia in human. TNF-α mainly inhibits the insulin induced tyrosine kinase activity of insulin receptor [13, 14]. So, it mainly reflects an acute phase response to myocardial attenuation of insulin receptor signaling through reducing auto-phosphorylation and tyrosine kinase activity [13].
TNF-α regulates expression of NF-κB which in turn induces expression of inflammatory and anti-apoptotic gene network [15]. However, TNF-α gene expression is a tightly regulated process. The -308 position of TNF-α promoter is a crucial one for binding of nuclear proteins and modulation of its expression [16]. Polymorphism at -308 position in TNF-α affect binding of transcription factors resulting elevated expression of TNF-α [17-21].
Kroeger et al. [17] reported that the G to A at -308 position resulted differential binding of an additional protein at -323 to -285 sequence. The binding of the extra protein alters the binding pattern of the common protein complex [17]. Another study [18] showed that the -308A polymorphic variant affects binding of transcription factor at -347 to -269 position of TNF promoter. Besides, the difference in TNF-α amount varies due to presence of several short tandem repeats such as TNFa and TNFc which are responsible for causing variation of TNF-α secretion by human monocytes [22]. However, it is clearly understood that serum TNF-α level might become a very important tool for determination of risk factor influencing atherosclerotic vascular lesions. Further, as TNF-α participates in numerous metabolic syndrome and TNF-α expression is varying due to allele differences at -308 position of promoter, it is being a putative nominee for CHD in the setting of chronic inflammatory disorder T2DM [23]. Therefore, we designed this study to determine association of TNF-α (-308G>A) polymorphism with CHD in T2DM patients in the northern region (Rajshahi) of Bangladesh.
MATERIALS AND METHODS
Recruitment of the study subject
This study included 96 T2DM patients who came in a diagnostic center from different places of Rajshahi, Bangladesh for routine test. Moreover, 10 more healthy participants having no family history of diabetes and CVD were included to use their genetic structure as wild type. A verbal consent was taken from all participants. We followed the guideline (Memo number: 58/320/IAMEBBC/IBSc) developed by the Institutional Animal, Medical Ethics, Biosafety and Biosecurity Committee (IAMEBBC), Institute of Biological Sciences, University of Rajshahi for experimentation on Animal, Human, Microbes and Living Natural Sources.
Blood collection and genomic DNA isolation
Blood of participants was collected in blood collection tube containing EDTAK3 (Qinghai Wuchan Trading Co. Ltd., China). After collection, blood samples were subjected in genomic DNA isolated using Genomic DNA isolation kit (Promega, USA) as per producer’s protocol. Then the DNA samples were purified using DNA purification kit (Promega, USA). Genomic DNA concentrations were equilibrated by ultra-violate spectrophotometry and agarose gel documentation. Salt removal of the purified genomic DNA was carried out by gel filtration using Sephacryl S-400 (GE Healthcare, USA).
High resolution melting (HRM) curve analysis and sequencing
For HRM analysis, we targeted a region of 110bp sequence in the TNF-α gene promoter. Before performing HRM, we optimized the PCR condition for specific HRM primers (Table 1) by gradient PCR [24] and the optimized condition were confirmed by normal PCR. The concentration of DNA was again normalized on the basis of qPCR Cq value. HRM was performed according to prior method [9] using GoTaq® qPCR master mix at 60°C for 40 cycle in Illumina Eco™ qPCR system (USA). The qPCR reaction mixture (10 µL) comprised of 5 µL (2x) GoTaq® qPCR master mix, 3 µL nuclease free water, 0.5 µL each of HRM primer, and 1 µL template DNA. qPCR was performed with the following cycling conditions: 95°C for 10 min, followed by 40 cycles at 95°C for 10 sec, 60°C for 30 sec, and 72°C for 15 sec. PCR specificity was confirmed by analyzing the melt curves at 95°C for 15 sec, 55°C for 15 sec, and 95°C for 15 sec. HRM data were processed and analyzed in Eco™ software integrated with the system. Identified genotypes were further verified after sequencing (1st BASE, Malaysia) corresponding samples. For sequencing, we used different primers set (Table 1) which were designed by targeting a 750 bp sequence from both directions.
Table 1. List of primers.
Statistical analysis
Fisher’s Exact test was performed to find out correlation between polymorphism and CVD. Kendall’s Tau correlation matrix was checked to evaluate the strength of correlation. Statistical analyses were performed using OPSTAT online software and SOCIOSTAT calculator.
RESULTS
HRM based polymorphism analysis and sequencing
Clear distinct melting curves were found between melting temperature (Tm) 81.5°C to 86.7°C. Melt curves that were similar in shape were distinguishable from each other by difference in Tm of the amplicon. In such situation, the Tm difference between samples was due to sequence variation from the wild type. Melt curves displaying a distinct shape from homozygote curves were usually due to the presence of base pairing mismatches (hetero duplexes) present in the PCR product mix. The magnitude of differences between samples is presented in Figure 1B. The melting curves of the ten healthy individuals were considered as wild type (GG) (Figure 1A). Curves from T2DM patients who have no mutation in any allele are aligned as wild type curve (Figure 1A and Figure 1B). Heterozygous mutated T2DM patient’s melting curves were shifting from wild type due to nucleotide variation in one of two alleles (Figure 1B) and gave two distinct Tm. On the other hand, patients having homozygous mutation displayed a similar pattern of melting curve with wild type due to nucleotide variation in two alleles (Figure 1B) and gave only one Tm. The difference in Tm was identified with the difference between fluorescence of each curve and wild type curve. Genotypes which were found between 84 and 85°C were grouped together. Different homozygotes showed similar shapes with a little difference in their Tm differing by 0.2–0.4°C. Although there were some variations (0.1–0.2°C) within each genotype, all homozygotes were clearly differentiated when the entire curves were considered. Heterozygotes displayed a different shape than that of homozygotes in the low melting region of transition (Figure 1B). The curves for 3 homozygous (A/A) mutations gave only one Tm which were 84.1, 83.9, and 83.9°C. On the other hand, the curve of 5 heterozygous (G/A) mutations gave Tm1 which were 83.8, 84°C, 84.1, 84.1°C, and 84.1°C while Tm2 were 79, 77.1, 79.1, 78.9, and 77.2°C respectively. However, mutations were identified and confirmed by comparing the pattern of our HRM melt curve with a well-established melt curve pattern reported previously [25]. After sequencing, we found sequence variation at -308 position of TNF-α gene (Figure 2) which is a heterozygous (G/A) mutation which was pre-confirmed in HRM analysis.
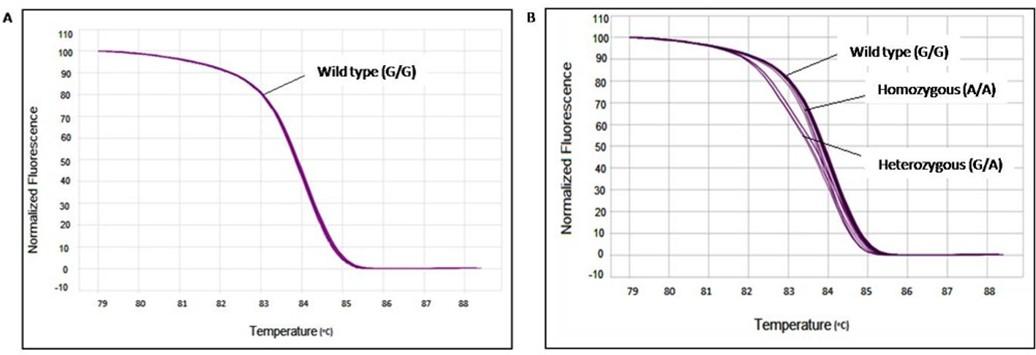
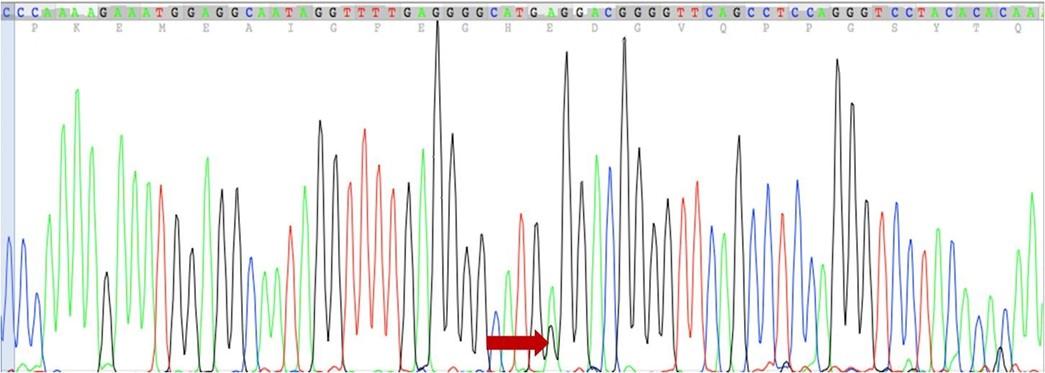
Correlation between TNF-α (-308G>A) polymorphism at promoter region and CHD
The result of HRM study showed three homozygous (A/A) and five heterozygous (G/A) mutations among tested 96 T2DM patients (Table 2). A total of 32 persons of all participants had CHD. Among the 32 CHD patients, six were bearing polymorphism (Table 3). The remaining two polymorphism carriers were not suffering from CHD though they were T2DM patients. However, according to the result of Fisher’s exact test, we found significant (P < 0.05) correlation between TNF-α polymorphism and CHD (Table 3). Besides, the Kendall’s Tau correlation matrix (r = 0.218) represents that there is a good relationship between TNF-α polymorphism and CHD (Table 4).
Table 2. Distribution of allele frequency of -308G>A SNP at TNF-α promoter region.
Table 3. Analysis of correlation between TNF-α (-308G>A) polymorphism at promoter region and CHD by Fisher’s Exact test.
Table 4. Analysis of strength of correlation between TNF-α (-308G>A) polymorphism at promoter region and CHD by Kendall’s Tau correlation matrix.
DISCUSSION
The frequency of CVD is being mounted at alarming figure in the developing countries, especially in Bangladesh where it is one of the major challenges for our health division. According to a most recent report published by World Health Organization (WHO) in 2018, the highest (30%) deaths were due to CVD among the total disease causing deaths [26]; while in 2014, only 17% deaths were of CVD [27]. The risk factors for CVD in Bangladeshi people are smoking, abdominal obesity, T2DM, hypertension, depression, high ApoB100/Apo-I ratio, physical inactivity, life style and peculiar food habit [28, 29]. Among these risk factors, T2DM is the only one which shares common risk factors with CVD [30]. T2DM is a disease which has two to four fold aptitude to induce CVD in people [31] indicating T2DM as an independent risk factor for CVD [32].
Beside, these risk factors, numerous common single-nucleotide polymorphisms (SNPs) are responsible for developing CVD and T2DM [33]. Such an example is paraoxonase which usually codes an enzyme that attaches with high-density lipoprotein, as well as protects low-density lipoprotein from proatherogenic oxidative alterations. But its polymorphic counterparts (Gln-Arg 192, or Met-Leu 54) guide to decrease enzymatic performance or decrease circulating enzyme content and are independently linked with both T2DM and CVD [34-37]. Further, there are some of the SNPs which are responsible for developing coronary artery disease (CAD) in T2DM patients. For example, adiponectin gene polymorphic variant (+276 G/T) has been showed to be linked with CAD in T2DM patients [38]. Again, gene that codes molecules concerned in the magnification of the inflammatory circumstance and proinflammatory cytokines could be an excellent nominee in determining risk in CAD [39]. However, inflammation is a very well acknowledged attribute of T2DM with elevated amount of proinflammatory cytokines, including IL-1, IL-6, and TNF-α [9]. A prior study [40] has established that the existence of the TNF-α -308A allele is linked with increased insulin resistance. Thus, based on the possible relationship of the insulin resistance syndrome with CAD, it is usually being hypothesized that the A variant represents the correlation between diabetes and amplified risk of CAD [39].
Interestingly, in this study, we are showing that TNF-α (-308G>A) polymorphism is associated with CHD in T2DM patients. We, the first group, are showing that TNF-α polymorphism is influencing CHD in T2DM patients in Bangladeshi people. A case-control study by Vendrell et al. [23] showed that TNF-α (-308G>A) polymorphism influences CHD in Caucasian T2DM patients. But the frequency of polymorphism (GA+AA = 40.6%) in Caucasian [23] is very high compared to the frequency (GA+AA = 8.33%) we observed in Bengali people. This noteworthy dissimilarity in term of frequency is perhaps due to the ethnic difference and environmental factors [41, 42]. A meta-analysis showed that TNF-α promoter (-308G>A) polymorphism has 1.5-fold amplified aptitude of influencing CHD in Caucasian [43]. Another case-control study [39] performed in Italian people stated that the incidence of TNF-α (-308G>A) polymorphism is significantly high in those people with diabetes.
However, in our another study [9], we reported association between TNF-α (-308G>A) polymorphism and T2DM in northern region of Bangladesh. Therefore, we can say that TNF-α (-308G>A) polymorphism is an important risk factor for developing CHD and T2DM in people from northern part of Bangladesh. Overall, this study may direct to conduct a further case-control study to evaluate the independent role of TNF-α (-308G>A) polymorphism in influencing CVD in Bangladeshi people.
ACKNOWLEDGEMENTS
The authors are thankful to the participants in this study for providing sample to perform this study. This research work did not receive any external financial support.
AUTHOR CONTRIBUTIONS
MRAM: Experimentation, data processing and analysis, manuscript writing and editing; MMH: Experimentation, data processing and analysis, manuscript writing and editing, manuscript drafting, manuscript revising and proof reading, DR: Experimentation, data processing and analysis; MSSU: Experimentation, data processing and analysis; BB: Experimentation, data processing and analysis; MAR: Conceptualization, data analysis, supervision; AH: Conceptualization, data analysis, supervision, resources.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Chu H, Yang J, Mi S, Bhuyan SS, Li J, Zhong L, Liu S, Tao Z, Li J, Chen H. Tumor necrosis factor-alpha G-308 A polymorphism and risk of coronary heart disease and myocardial infarction: a case–control study and meta-analysis. Journal of Cardiovascular Disease Research. 2012; 3:84-90.
- [2]Asifa GZ, Liaquat A, Murtaza I, Kazmi SA, Javed Q. Tumor necrosis factor-alpha gene promoter region polymorphism and the risk of coronary heart disease. The Scientific World Journal. 2013; 1:2013.
- [3]Morange PE, Tregouet DA, Godefroy T, Saut N, Bickel C, Rupprecht HJ, Lackner K, Barbaux S, Poirier O, Peiretti F, Nalbone G. Polymorphisms of the tumor necrosis factor-alpha (TNF) and the TNF-alpha converting enzyme (TACE/ADAM17) genes in relation to cardiovascular mortality: the AtheroGene Journal of Molecular Medicine. 2008; 86:1153-1161.
- [4]Szekanecz Z. Pro-inflammatory cytokines in atherosclerosis. Israel Medical Association Journal. 2008; 10:529-530.
- [5]Cesari M, Penninx BW, Newman AB, Kritchevsky SB, Nicklas BJ, Sutton-Tyrrell K, Rubin SM, Ding J, Simonsick EM, Harris TB, Pahor M. Inflammatory markers and onset of cardiovascular events: results from the Health ABC study. Circulation. 2003; 108:2317-2322.
- [6]Hajeer AH, Hutchinson IV. Influence of TNFα gene polymorphisms on TNFα production and disease. Human Immunology. 2001; 62:1191-1199.
- [7]Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature. 1997; 385:729-733.
- [8]Moller DE. Potential role of TNF-α in the pathogenesis of insulin resistance and type 2 diabetes. Trends in Endocrinology & Metabolism. 2000; 11:212-217.
- [9]Roy D, Ullah MS, Basak B, Hasan MM, Tanzim M, Haque A. Identification of TNF-α -308G/A (rsl800629) polymorphism in Bangladeshi population related to type-2 diabetes. Journal of Applied Biology & Biotechnology. 2019; 7(06):25-30.
- [10]Zhang H, Park Y, Wu J, Lee S, Yang J, Dellsperger KC, Zhang C. Role of TNF-α in vascular dysfunction. Clinical Science. 2009; 116:219-230.
- [11]Skoog T, Dichtl W, Boquist S, Skoglund-Andersson C, Karpe F, Tang R, Bond MG, De Faire U, Nilsson J, Eriksson P, Hamsten A. Plasma tumour necrosis factor-α and early carotid atherosclerosis in healthy middle-aged men. European Heart Journal. 2002; 23:376-383.
- [12]Heijmans BT, Westendorp RG, Droog S, Kluft C, Knook DL, Slagboom PE. Association of the tumour necrosis factor α −308G/A polymorphism with the risk of diabetes in an elderly population-based cohort. Genes & Immunity. 2002; 3:225-228.
- [13]Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-α -and obesity-induced insulin resistance. Science. 1996; 271(5249):665-670.
- [14]Hotamisligil GS. The role of TNFα and TNF receptors in obesity and insulin resistance. Journal of Internal Medicine. 1999; 245:621-625.
- [15]Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-κB control. BMC Genomics. 2005; 6:137.
- [16]Kroeger KM, Abraham LJ. Identification of an AP-2 element in the -323 to -285 region of the TNF-α gene. Biochemistry and Molecular Biology International. 1996; 40:43–51.
- [17]Kroeger KM., Carville KS, Abraham LJ. The -308 tumor necrosis factor-a promoter polymorphism effects transcription. Molecular Immunology. 1997; 34:391–399.
- [18]Wu WS, McClain KL. DNA polymorphisms and mutations of the tumor necrosis factor-α (TNF-α) promoter in Langerhans cell histiocytosis (LCH). Journal of Interferon & Cytokine Research. 1997; 17:631-635.
- [19]Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor α promoter on transcriptional activation. Proceedings of the National Academy of Sciences. 1997; 94:3195-3199.
- [20]Groote DE. Tumour necrosis factor (TNF) gene polymorphism influences TNF‐α production in lipopolysaccharide (LPS) ‐stimulated whole blood cell culture in healthy humans. Clinical & Experimental Immunology. 1998; 113:401-406.
- [21]Ghazouani L, Abboud N, Addad F, Khalfallah AB, Brahim N, Mediouni M, Almawi WY, Mahjoub T. -308G>A and 1031T>C tumor necrosis factor gene polymorphisms in Tunisian patients with coronary artery disease. Clinical Chemistry and Laboratory Medicine (CCLM). 2009; 47:1247-1251.
- [22]Pociot F, Briant L, Jongeneel CV, Mölvig J, Worsaae H, Abbal M, Thomsen M, Nerup J, Cambon‐Thomsen A. Association of tumor necrosis factor (TNF) and class II major histocompatibility complex alleles with the secretion of TNF‐α and TNF‐β by human mononuclear cells: a possible link to insulin‐dependent diabetes mellitus. European Journal of Immunology. 1993; 23:224-231.
- [23]Vendrell J, Fernandez-Real JM, Gutierrez C, Zamora A, Simon I, Bardaji A, Ricart W, Richart C. A polymorphism in the promoter of the tumor necrosis factor-α gene (− 308) is associated with coronary heart disease in type 2 diabetic patients. Atherosclerosis. 2003; 167:257-264.
- [24]Roy D, Hasan MM, Haque A. Mutation detection sensitivity of high resolution melting in clinical samples: a comparative study between formamide and dimethyl sulfoxide. Journal of Advanced Biotechnology and Experimental Therapeutics. 2019; 2(2):51-54.
- [25]You CG, Li XJ, Li YM, Wang LP, Li FF, Guo XL, Gao LN. Association analysis of single nucleotide polymorphisms of proinflammatory cytokine and their receptors genes with rheumatoid arthritis in northwest Chinese Han population. Cytokine. 2013; 61:133-138.
- [26]World Health Organization. Noncommunicable Diseases (NCD), Country Profiles 2018.Available at: https://www.who.int/nmh/countries/2018/bgd_en.pdf?ua=1
- [27]World Health Organization. Noncommunicable Diseases (NCD), Country Profiles 2014. Available at: https://www.who.int/nmh/countries/2014/bgd_en.pdf?ua=1
- [28]Saquib N, Saquib J, Ahmed T, Khanam MA, Cullen MR. Cardiovascular diseases and type 2 diabetes in Bangladesh: a systematic review and meta-analysis of studies between 1995 and 2010. BMC Public Health. 2012; 12:434.
- [29]Islam AM, Mohibullah AK, Paul T. Cardiovascular disease in Bangladesh: a review. Bangladesh Heart Journal. 2016; 31:80-99.
- [30]De Rosa S, Arcidiacono B, Chiefari E, Brunetti A, Indolfi C, Foti DP. Type 2 diabetes mellitus and cardiovascular disease: genetic and epigenetic links. Frontiers in Endocrinology. 2018; 9:2.
- [31]Kannel WB, McGee DL. Diabetes and cardiovascular disease: the Framingham study. JAMA. 1979; 241:2035-2038.
- [32]Grundy SM, Benjamin IJ, Burke GL, Chait A, Eckel RH, Howard BV, Mitch W, Smith Jr SC, Sowers JR. Diabetes and cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation. 1999; 100:1134-1146.
- [33]Ma RC. Genetics of cardiovascular and renal complications in diabetes. Journal of Diabetes Investigation. 2016; 7:139-154.
- [34]Ruiz J, Morabia A, Blanche H, James RW, Garin MB, Charpentier G, Passa P, Vaisse C, Froguel P. Gln-Arg192 polymorphism of paraoxonase and coronary heart disease in type 2 diabetes. The Lancet. 1995; 346:869-872.
- [35]Serrato M, Marian AJ. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. Journal of Clinical Investigation. 1995; 96:3005-3008.
- [36]Blatter Garin MC. Paraoxonase polymorphism Met-Leu 54 is associated with modified serum concentrations of the enzyme. Journal of Clinical Investigation. 1997; 99:62-66.
- [37]Pfohl M, Koch M, Enderle MD, Kühn R, Füllhase J, Karsch KR, Häring HU. Paraoxonase 192 Gln/Arg gene polymorphism, coronary artery disease, and myocardial infarction in type 2 diabetes. Diabetes. 1999; 48:623-627.
- [38]Bacci S, Menzaghi C, Ercolino T, Ma X, Rauseo A, Salvemini L, Vigna C, Fanelli R, Di Mario U, Doria A, Trischitta V. The +276 G/T single nucleotide polymorphism of the adiponectin gene is associated with coronary artery disease in type 2 diabetic patients. Diabetes Care. 2004; 27:2015-2020.
- [39]Sbarsi I, Falcone C, Boiocchi C, Campo I, Zorzetto M, De Silvestri A, Cuccia M. Inflammation and atherosclerosis: the role of TNF and TNF receptors polymorphisms in coronary artery disease. International Journal of Immunopathology and Pharmacology. 2007; 20:145-154.
- [40]Fernandez-Real JM, Gutierrez C, Ricart W, Casamitjana R, Fernandez-Castaner M, Vendrell J, Richart C, Soler J. The TNF-α Gene Neo I polymorphism influences the relationship among insulin resistance, percent body fat, and increased serum leptin levels. Diabetes. 1997; 46:1468-1472.
- [41]Haghani K, Bakhtiyari S. The study on the relationship between IRS-1 Gly972Arg and IRS-2 Gly1057Asp polymorphisms and type 2 diabetes in the Kurdish ethnic group in West Iran. Genetic Testing and Molecular Biomarkers. 2012; 16:1270-1276.
- [42]Saberi H, Mohammadtaghvaei N, Gulkho S, Bakhtiyari S, Mohammadi M, Hanachi P, Gerayesh-Nejad S, Zargari M, Ataei F, Parvaneh L, Larijani B. The ENPP1 K121Q polymorphism is not associated with type 2 diabetes and related metabolic traits in an Iranian population. Molecular and Cellular Biochemistry. 2011; 350:113-118.
- [43]Zhang HF, Xie SL, Wang JF, Chen YX, Wang Y, Huang TC. Tumor necrosis factor-alpha G-308A gene polymorphism and coronary heart disease susceptibility: an updated meta-analysis. Thrombosis Research. 2011; 127:400-405.