Aberrant methylation of CDKN2A, RASSF1A and WIF1 in sporadic adenocarcinomatous colorectal cancer: Associations with clinicopathological features
Abstract
Accumulating evidence support that aberrant methylation of various cancer-related genes plays an important role in the initiation and progression of colorectal cancer (CRC). This study aims to validate the accuracy of methylation specific polymerase chain reaction (MSP) to assess frequency and distribution of GSTP1, CDKN2A, RASSF1A, and WIF1 methylation and analyse their correlation with clinicopathological variables in sporadic adenocarcinomatous CRC. Of the 248 CRC tissues, methylation was identified in 7.7% for GSTP1, 22.2% for CDKN2A, 33.1% for RASSF1A, and 54.4% for WIF1. Hypermethylation of CDKN2A, RASSF1A, and WIF1 was significantly associated with adenocarcinoma (p< 0.001), mucinous adenocarcinoma (p< 0.001), and signet-ring cell adenocarcinoma subtypes (p = 0.017), respectively. Both CDKN2A and WIF1 methylations were more common in stage II (p = 0.012 for CDKN2A and p = 0.010 for WIF1) and absence of lymph node metastasis (p = 0.011 for CDKN2A and p = 0.012 for WIF1) but were less common in stage III (p = 0.016 for CDKN2A and p = 0.010 for WIF1). RASSF1A methylation was associated with moderate differentiation (p = 0.038). These findings suggest that methylation of CDKN2A, RASSF1A, and WIF1 may significantly contribute to CRC pathogenesis and may be considered as valuable biomarkers for accessing the development and progression of particular subtypes of colorectal cancer.
INTRODUCTION
Colorectal cancer (CRC) is a common malignant cancer as well as a leading cause of cancer mortality worldwide, and still has poor prognosis. Although the exact pathologic mechanism has not been understood fully, it is widely accepted that CRC development is resulted from the accumulation of multiple genetic and epigenetic alterations [1]. DNA promoter methylation, one of the main mechanisms of epigenetic modifications, is to be associated with development and progression of human cancers [1, 2]. Aberrant DNA promoter methylation, which is characterized by covalent addition of a methyl group to the 5’ position on Cytosine residues of CpG islands, often occurs in the earliest precursor lesion (aberrant crypt foci), and in the early stage of colorectal carcinogenesis [3, 4]. Promoter CpG island DNA hypermethylation of cancer-related genes leads to transcriptional gene silencing and importantly contributes to colorectal tumorigenesis [5].
Cyclin dependent kinase inhibitor 2A (CDKN2A), Ras association domain family 1 isoform A (RASSF1A), and Wnt inhibitory factor 1 (WIF1) genes function as important tumor suppressors, and their activation results in cell cycle arrest, senescence, and apoptosis [2, 6, 7]. Glutathione S-transferase pi 1(GSTP1) is proposed to act as a “caretaker” gene that detoxifies reactive electrophilic intermediates/carcinogenic compounds [8]. GSTP1, CDKN2A, RASSF1A, and WIF1 promoter methylations are frequent epigenetic events in various human cancers, including CRC, and crucial mechanisms leading to cell overgrowth, uncontrolled cell proliferation, tumor development and progression [6, 8, 9].
Although GSTP1, CDKN2A, RASSF1A, and WIF1 inactivation by aberrant DNA methylation has been widely studied in CRC, associations between GSTP1, CDKN2A, RASSF1A, or WIF1 and clinicopathological features of CRC remain controversial. Therefore, the present study was conducted to elucidate the frequency of GSTP1, CDKN2A, RASSF1A, and WIF1 methylation and the correlation of each with clinicopathological data.
MATERIALS AND METHODS
Patients and tissue specimens
A total of 248 tumors of sporadic adenocarcinomatous CRC were collected for analysis in the present study. Clinical data of the patients were collected from the hospital records. Written consent was obtained from all patients was approved by the Ethnic Committee of National Cancer Hospital K (Circular No.04/2008/TT-BYT). Diagnostic pathology were evaluated by more than two pathologists based on the World Health Organization (WHO) classification (WHO, 2019) guidelines.
DNA extraction and bisulfite modification
Genomic DNA was extracted from 5 sections of 10 μm thickness of macro-dissected colorectal tumor tissues using QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA) (containing at least 30% tumor cells). To evaluate the quality of DNA specimens, Polymerase Chain Reaction (PCR) for single-copy gene β-globin was carried out. DNA samples were then introduced to sodium bisulfite conversion using EpiTect Bisulfite Kit (Qiagen, Valencia, CA, USA).
Methylation specific polymerase chain reaction (MSP)
For each sample, methylation status of GSTP1, CDKN2A, RASSF1A, and WIF1 were evaluated by using methylation specific polymerase chain (MSP). Sodium bisulfite-treated DNA samples were used as templates for PCR with specific primers, which were designed to be specific to either the methylated or unmethylated sequence of each gene. The reluctant PCR products were separated on 10% Poly-acrylamide gel. Each MSP was performed at least twice. Primer sequences for each gene are listed in the Table 1.
Table 1. Primer sequences.
Statistical analysis
Statistical analysis was performed using SPSS software (IBM Corporation, New York, NY, USA). Fisher’s exact test or χ2 test was used to determine the association of variables properly. A p-value less than 0.05 (typically ≤ 0.05) is statistically significant.
RESULTS
Patient characteristics
Table 2 summarizes clinicopathological characteristics of 248 patients. The median age at diagnosis was 60 years (range, 26-90 years). Histological analysis revealed 75.4% adenocarcinomas, 21.8% mucinous adenocarcinomas, and 2.8% signet ring cell adenocarcinomas. Most tumors were moderately differentiated (64.5%), and there were only 4.8% of tumors being well differentiated and 6.0% poorly differentiated (excepting for 61 cases without tumor differentiation evaluation). The differentiation criteria used in this study according to the WHO’s classification [10]. The majority of patients (91.9%) had local disease at initial diagnosis (Table 2).
Table 2. Clinicopathological characteristics of the patients with CRC.
Associations between GSTP1, CDKN2A, RASSF1A, or WIF1 and clinicopathological features of CRC
Aberrant promoter methylation of GSTP1, CDKN2A, RASSF1A, and WIF1 was detected in 19 (7.7%), 55 (22.2%), 82 (33.1%), and 135 (54.4%) in a total of 248 colorectal tumors, respectively (Figure 1). GSTP1 methylation tended to be associated with male patients (p = 0.053) and moderate tumor differentiation (p = 0.082), yet GSTP1 hypermethylation did not significantly correlate with any clinicopathological feature. CDKN2A methylation was more common in adenocarcinoma (p< 0.001) but less common in mucinous adenocarcinoma (p< 0.001); in contrast, RASSF1A methylation was more frequent in mucinous adenocarcinoma (p< 0.001) but less frequent in adenocarcinoma (p = 0.002). Aberrant promoter methylation of WIF1 occurred frequently in signet-ring cell adenocarcinoma (p =0.017) but rarely in mucinous adenocarcinoma (p = 0.009). A statistically significant correlation between methylation and pathologic stage was observed, where both CDKN2A and WIF1 methylations were more common in stage II (p = 0.012 for CDKN2A and p = 0.010 for WIF1) and less common in stage III (p = 0.016 for CDKN2A and p = 0.011 for WIF1). Moreover, CDKN2A and WIF1 methylations were associated with the absence of lymph node metastasis (p = 0.014 and p = 0.012, respectively). RASSF1A hypermethylation significantly correlated with moderate tumor differentiation (p = 0.038) and had tendencies to be less common in stage III (p = 0.056) and in lymph node metastasis (p = 0.072) (Table 3).
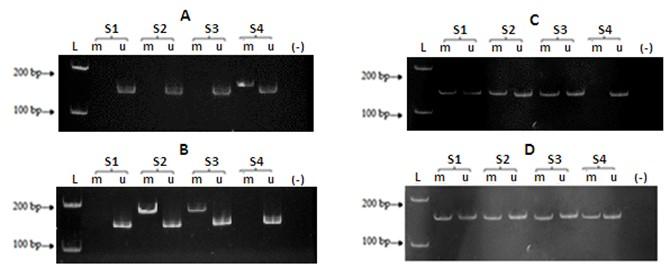
Table 3. GSTP1, CDKN2A, RASSF1A, and WIF1 methylations and correlations with clinicopathological features.
DISCUSSION
Identifying molecular abnormalities has been not only essential to understand the pathogenesis of the disease better, but also valuable in diagnosis, prognosis, selection of optimal therapeutic regimens, and discovery of risk factors associated with a particular subtype [1]. Multiple studies on methylation of tumor suppressor genes, such as CDKN2A, RASSF1A, and WIF1, have been reported in CRC; however, their results are inconsistent. In fact, epigenetic patterns are modulated by both endogenous and exogenous factors, including aging, ethnicity, gender, dietary habits, lifestyles, environmental factors, and medications [11, 12]. This study showed the frequency and relationship of GSTP1, CDKN2A, RASSF1A, and WIF1 methylation with clinicopathological features specific to the Vietnamese CRC population.
In the present study, GSTP1, CDKN2A, RASSF1A, and WIF1 promoter methylation was found in 7.7%, 22.2%, 33.1%, and 54.5% of CRC tumors, respectively. This shows that WIF1 is a commonly methylated gene, while GSTP1 methylation seems to be a rare event in Vietnamese CRC patients. Through extensively screening reports into CRC, the rate of CDKN2A (22.2%) and RASSF1A methylation (33.1%) was approximately the same average frequency as reported in meta-analyses [6, 7]. Whereas, our methylation frequency of 54.5% for WIF1 is slightly lower than that in an earlier literature, which indicated frequency as high as 80.6% [13]. Generally, the frequency of WIF1 methylation has been found to be relatively high in CRC [14], suggesting that WIF1 hypermethylation is a frequent event in CRC. Considering the differences in genetic and environmental factors related to CRC, it is possible that prevalence of epigenetic alterations varies among studied population. Recent evidence has shown that DNA methylation is incompatible in distinct races and ethnicities [15].
Our study revealed that CDKN2A methylation frequently occurred in adenocarcinoma but rarely in mucinous adenocarcinoma, whereas RASSF1A methylation was more common in mucinous adenocarcinoma but less common in adenocarcinoma. Frequency of WIF1 hypermethylation was positively associated with singlet-ring cell adenocarcinoma and inversely associated with mucinous adenocarcinoma. Although correlation between CDKN2A, RASSF1A, or WIF1 methylation and histologic subtypes remains unknown, our previous study also showed a significant association between RASSF1A hypermethylation and mucinous adenocarcinoma in CRC [16]. These observations clearly indicated that CDKN2A, RASSF1A, and WIF1 methylation targets different histologic subtypes of CRC. However, this study is limited by the small sample size in histologic subtypes. Thus, further studies with a larger sample size are essential to confirm this hypothesis.
Our analysis showed that CDKN2A hypermethylation was significantly associated with several clinicopathological characteristics toward a good prognosis. CDKN2A promoter methylation was found frequently in cases with early-stage and absence of lymph node metastasis. These results suggest that CDKN2A methylation plays a crucial role in the initiation of CRC. In contrast, several reports showed that CDKN2A promoter hypermethylation frequently occurred in more malignant CRC phenotype, which was associated with advanced stage and lymph node metastasis [6]. This discrepancy may be attributed to sample size, sample selection, and method used.
Similar to CDKN2A methylation, aberrant methylation of WIF1 was significantly associated with tumor stage, in which WIF1 hypermethylation frequently occurred in stage II but rarely in stage III. In addition, WIF1 promoter methylation was found commonly in cases without lymph node metastasis. These results are consistent with a previous study, which showed a relatively high frequency of WIF1 methylation (up to 74%) in patients with stage I and II sporadic CRC compared with 2% in healthy individuals [14]. The increased level of WIF1 methylation and the down-regulation of WIF1 expression have been observed in colorectal adenoma tissues [17, 18]. Based on these observations, aberrant promoter methylation of WIF1 may be related to tumor initiation.
Methylation status of RASSF1A was obviously correlated with moderate differentiation, which is consistent with a previous report [15]. Correlation between RASSF1A methylation and pathologic stage varies across various studies; some reports observed a higher level of RASSF1A methylation in early-stage of CRC while others reported more frequent RASSF1A methylation on later-stage [19, 20]. Although not significant, RASSF1A hypermethylation was found rarely in stage III CRC and lymph node metastasis in the present study.
In conclusion, this study reports presence of GSTP1, CDKN2A, RASSF1A, and WIF1 methylation in the Vietnamese CRC population, and their correlations with clinicopathological characteristics. These observations suggest that aberrant methylation of CDKN2A, RASSF1A, and WIF1 may be related to tumor initiation but not to tumor progression. CDKN2A, RASSF1A, and WIF1 methylations are considered as valuable diagnostic and prognostic markers in accessing the development and progression of particular subtypes of colorectal cancer.
ACKNOWLEDGEMENT
The authors received no financial support for this project.
AUTHOR CONTRIBUTIONS
L.D.V. and Q.N.N.: Conception and Design of the experiments. H.V.N.: Methodology and Data analysis, V-.L.T.: Data curation and Writing – original draft, L.D.V: Writing – review and editing. Q.N.N.: Supervision. All authors reviewed the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome research. 2012;22:271-282.
- [2]Hu H, Li B, Zhou C, Ying X, Chen M, Huang T, et al. Diagnostic value of WIF1 methylation for colorectal cancer: a meta-analysis. Oncotarget. 2018;9:5378-5386.
- [3]Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. The American journal of pathology. 2002;160:1823-1830.
- [4]Ashktorab H, Brim H. DNA Methylation and Colorectal Cancer. Current colorectal cancer reports. 2014;10:425-30.
- [5]Patai ÁV, Molnár B, Kalmár A, Schöller A, Tóth K, Tulassay Z. Role of DNA Methylation in Colorectal Carcinogenesis. Digestive Diseases. 2012;30:310-315.
- [6]Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J, et al. The prognostic value of CDKN2A hypermethylation in colorectal cancer: a meta-analysis. British journal of cancer. 2013;108:2542-2548.
- [7]Hu H, Zhou C, Li B, Chen Y, Dai J, Mao Y, et al. Diagnostic value of RASSF1A hypermethylation in colorectal cancer: a meta-analysis. Pathology, research and practice. 2018;214:1572-1578.
- [8]Meiers I. Glutathione S-Transferase pi (GSTP1). Atlas Genet Cytogenet Oncol Haematol. 2010;14:1181-5.
- [9]Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837-845.
- [10]Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182-188.
- [11]Lao VV, Grady WM. Epigenetics and colorectal cancer. Nature reviews Gastroenterology & hepatology. 2011;8:686-700.
- [12]Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nature reviews Genetics. 2012;13:679-692.
- [13]Yamaoka S, Yamamoto H, Nosho K, Taniguchi H, Adachi Y, Sasaki S, et al. Genetic and epigenetic characteristics of gastric cancers with JC virus T-antigen. World journal of gastroenterology. 2009;15:5579-5585.
- [14]Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6185-6191.
- [15]Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623-629.
- [16]Ta TV, Nguyen QN, Chu HH, Truong VL, Vuong LD. RAS/RAF mutations and their associations with epigenetic alterations for distinct pathways in Vietnamese colorectal cancer. Pathology, research and practice. 2020;216:152898.
- [17]Taniguchi H, Yamamoto H, Hirata T, Miyamoto N, Oki M, Nosho K, et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946-7952.
- [18]Belshaw NJ, Elliott GO, Foxall RJ, Dainty JR, Pal N, Coupe A, et al. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. British journal of cancer. 2008;99:136-142.
- [19]Fernandes MS, Carneiro F, Oliveira C, Seruca R. Colorectal cancer and RASSF family–a special emphasis on RASSF1A. International journal of cancer. 2013;132:251-258.
- [20]Coppedè F, Migheli F, Lopomo A, Failli A, Legitimo A, Consolini R, et al. Gene promoter methylation in colorectal cancer and healthy adjacent mucosa specimens: correlation with physiological and pathological characteristics, and with biomarkers of one-carbon metabolism. Epigenetics. 2014;9:621-633.