No evidence of relationship between colorectal cancer susceptibility and ERCC2 gene polymorphisms
Abstract
Excision repair cross-complementing group 2 gene (ERCC2) polymorphisms have been linked as being a risk factor for colorectal cancer (CRC) emergence. However, data from several studies are contradictory. To validate genetic biomarkers of the CRC, the impact of the following ERCC2 polymorphism (rs1799793 and rs238406) was examined on CRC susceptibility among sample of Iraqi population. A total of 126 subjects were enrolled in this case control study; 78 CRC patients and 48 apparently healthy individuals who are matched for age, gender, smoking status, and BMI. Polymerase chain reaction (PCR) was used for genotyping, followed by sequencing then the association between genetic polymorphisms and CRC risk was investigated. No associations were detected between ERCC2 genotypes or haplotypes and CRC susceptibility. Even though there was strong linkage disequilibrium (D′=0.82). After stratification according to demographics of the participants, no effects were observed for age, gender, smoking status, and body mass index (BMI). Taken together, the results suggest that ERCC2 polymorphisms do not influence CRC development.
INTRODUCTION
Deoxyribonucleic acid (DNA) damage repair is crucial to avoid many diseases like and not limited to malignancies [1]. Individuals’ ability to repair DNA varies greatly, as a large number of studies have shown that persons with lower DNA repair capacity are highly likely to acquire cancer [2]. One of the important genetic mutations in DNA repair genes that have been considered to increase the individual’s carcinogenesis susceptibility is excision-repair cross complementing group 2 (ERCC2) variants [3]. ERCC2 encodes an ATP-dependent DNA helicase enzyme responsible for DNA unwinding in the 5’-3’ direction. Its role is pivotal in eliminating helix-distorting base lesions resulting from endogenous and exogenous factors. ERCC2 gene is located on chromosome19q13.3, composed of 23 exons and has more than 100 variants that have been reported in earlier research [4, 5]. Of the investigated single nucleotide polymorphisms (SNPs) are missense mutation of Guanine to Adenine substitution in exon 10 leading to an aspartic acid (Asp) to asparagine (Asn) substitution in the codon 312 (rs1799793) and a synonymous Cytosine to Adenine substitution in exon 6 though keeping the arginine amino acid in the codon 156 (rs238406) [3]. As ERCC2 is a critical gene for nucleotide excision repair pathway (NER), the association between ERCC2 polymorphisms and cancer susceptibility is of particular interest.
Colorectal cancer (CRC) is the third most common cancer globally, and according to the International Agency for Research on Cancer (IARC), CRC incidence will rise by 56% in the next 20 years, to more than 3 million new cases per year. Furthermore, the approximate growth in the number of deaths may even be higher by 69%, to about 1.6 million deaths globally in 2040 [6, 7]. In Iraq, CRC is also the third most common cancer with a percentage of 6.97 detected in the year 2020 and cases showed a significant surge after the year 2007 similarly to other developing countries [8, 9]. In addition, many cases are reported in younger individuals in Iraq, suggesting that this malignancy requires more attention from health care system [10]. There are several risk factors that have been studied extensively and thought to be a major contributor to CRC development [11]. Concerning molecular features, CRC has three phenotypes: microsatellite instability, chromosomal instability and cytosine guanine (CpG) island methylator phenotype [12, 13]. Aberrant methylation of some genes (cyclin dependent kinase inhibitor 2A, Ras association domain family 1 isoform A, Wnt inhibitory factor 1 and glutathione S-transferase has been investigated as a possible link not only in CRC pathogenesis but also affecting CRC clinicopathologic characteristics such as histologic subtype and CRC stage [14]. Risk of CRC development was also examined with other genes that are involved with EGFR signal transduction pathway and found to have an impact on clinicopathologic parameters as well [15].
Emerging evidence indicates that the DNA repair defects are involved in the initiation and worsening of CRC [16, 17]. Therefore, several research have investigated the relationship of DNA repair gene polymorphisms, particularly ERCC2 rs238406 and rs1799793. Nevertheless, the findings are inconclusive where Zhang et al. concluded that rs238406 has no impact on CRC susceptibility in Chinese population [18]. On the other hand, Swedish study, and Polish research both found that this SNP is associated with increased CRC risk [19, 20]. Regarding Asp 312 Asn variant, the same Polish study found that individuals with variant genotype are more prone to develop CRC compared to wild type carriers [20]. In 2014, a study conducted to examine the relationship between diet, genetic polymorphisms of several genes including ERCC2 Asp 312 Asn and CRC risk and authors found carriers of GG genotype had protective effect against CRC risk while AG and AA genotypes had lower DNA repairing capacity and elevated the risk by 1.84 times [21]. However, a study conducted in Tiwan in 2016 on 362 CRC patients who were matched with age and gender to 362 healthy subjects found lack of association with rs1799793 [22]. Moreover, Gil et al. examined the genes in nucleotide excision repair and base excision repair pathways where Asp 312 Asn variant did not reach statistical significance [23]. Accordingly, the aim of this study is to investigate the relationship of the ERCC2 SNPs mentioned above with CRC susceptibility among a sample of Iraqi population.
MATERIALS AND METHODS
Study subjects
This hospital-based case-control study involved 78 CRC patients and 48 disease-free control subjects whose age, gender, smoking-status, and Body Mass Index (BMI) are matched. All cases were recruited from Oncology Teaching Hospital-Medical city/ Baghdad from February to the end of December 2022. A structured questionnaire was used to collect demographic such as age, gender, wight, height and smoking status from study participants. Meanwhile, clinical data such as CRC stage, tumor location tumor invasion, lymph nodes involvement and the presence of metastasis were obtained from patient’s medical file. The research was approved by the ethics committee in College of Pharmacy, University of Baghdad (approval number: RECAUBCP26102021B) in agreement with the ethical standards of the responsible committee on human experimentation (institutional and national). Informed consent was acquired from all participants before inclusion into the study. CRC diagnosis was confirmed by colonoscopy and a histopathological biopsy [24]. CRC was staged according to TNM (Tumor Node Metastasis) classification system [25]. Inclusion criteria involved patients with confirmed CRC diagnosis who are over the age of 18 years. Exclusion criteria were patients who have or had other malignancies and patients with neurological or genetic disease.
DNA extraction and genotyping
Peripheral venous blood samples of three milliliters were taken from CRC patients and CRC free controls in K3EDTA coated tubes. DNA extraction was accomplished by ReliaPrep™ Blood gDNA Miniprep System (Promega, USA) according to manufacturer’s instructions. Quantus Fluorometer was utilized to detect the concentration and quality of the DNA extracted. DNA sequence to be amplified was obtained from the national center for biotechnology information (NCBI) GenBank database.
ERCC2 primers were designed using Premier 3 software (version 0.4.0). Data regarding primers’ annealing temperature, primers length, Polymerase chain reaction (PCR) amplicon length, and primer sequence are found in Table 1. PCR technique was operated using a thermal cycler (Thermo Fisher Scientific, USA). PCR products were detected using Agarose gel electrophoresis to confirm the presence of amplification. Afterwards, PCR amplicons were sent to Macrogen Corporation – Korea for Sanger sequencing using automated DNA sequencer then the results were analyzed by genius software V 2021.1.1) (Biomatters Ltd., Auckland, New Zealand; www.geneious.com).
Table 1. Primers are used in the amplification process.
Statistical analysis
Data were analyzed using SPSS software version 26 (SPSS® Inc, Chicago, USA). Discrete variables were expressed as count and percentage while Continuous data were expressed as mean ± Standard deviation (SD). The relation between variables were assessed by Chi-square or Fisher exact test as appropriate. p value < 0.05 was statistically significant.
Estimation of haplotypes between ERCC2 SNPs as well as linkage disequilibrium was done by SHEsis online software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci [26]. Linkage disequilibrium was expressed by Lewontin’s coefficient (D’).
RESULTS
Participants characteristics
The present study comprised a total of 126 subjects (78 CRC patients and 48 apparently healthy individuals). Demographic and clinical data of the participants are illustrated in Table 2. The average age of patients and healthy controls was (55.4 ± 12.5 and 53.1 ± 17.04) years, respectively. Significant difference in mean age distributions between CRC and disease-free groups was lacking (P value= 0.42). Male gender constituted 56.4% of CRC patients and 52.1% of healthy group (P value=0.82). There was no significant difference between cases and control groups with respect to smoking status and BMI (P value= 0.65; 0.23, respectively). Most of the patients had their tumor in the colon (65.4%) and most of these tumors were moderately differentiated (62.8%).
Table 2. Characteristics of patients with colorectal cancer and controls.
Genotype, haplotype, and allele distribution and its association with CRC risk
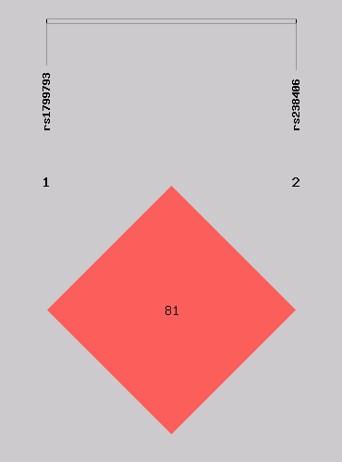
Within the control and CRC groups under research, both evaluated SNPs were in accordance with Hardy-Weinberg Equilibrium. This suggests that any differences in the study population are due to variations attributed to SNPs that have significantly different frequencies in the control group and the population with colorectal cancer in the present study. However, no association between CRC patients and CRC free controls was found for the aforementioned SNPs in codominant, recessive and dominant models (Table 3 and 4). There was strong linkage disequilibrium between the two SNPs (D'=0.83) as the distance between the two SNPs is 1050 bp (Figure 1). Four different haplotypes appeared in our analysis. The most frequent haplotype was A_C [cases: 63.38 (41.2%) vs. control: 36.82 (43.8%)] and the least frequent haplotype was A_A [cases: 6.62 (4.3%) vs. control: 1.18 (1.4%)] in both groups. However, there were no differences between patients and controls in the overall or individual haplotype distributions (Global test, P value=0.322) (Table 5).
Table 3. Genotypes and allele frequencies for rs238406 in colorectal cancer patients and controls.
Table 4. Genotypes and allele frequencies for rs1799793 in colorectal cancer patients and controls.
Table 5. Haplotype distribution of ERCC2 SNPs in colorectal cancer patients and controls.
Stratification analysis by demographics and clinicopathological characteristics for ERCC2 SNPs and CRC risk
No statistical significance was found between ERCC2 genotypes and tumor location, tumor differentiation, CRC stage, tumor invasion, lymph node involvement or metastasis (Table 6). The possible relationship between ERCC2 genetic variants and the risk of CRC development was further analyzed through stratification by age, gender, smoking status, and BMI. Nevertheless, no association was found between rs1799793 or rs238406 and study participants’ characteristics (Table 7 and 8).
Table 6. Association between ERCC2 polymorphisms and clinicopathological features of CRC.
Table 7. Association between rs238406 and risk of CRC stratified by demographic characteristics.
Table 8. Association between rs1799793 and risk of CRC stratified by demographic characteristics.
DISCUSSION
CRC is a multi-step process that begins with an aberrant polyp growth that comes from the colon's innermost layer. A polyp may take 10 to 15 years to turn into a cancerous tumor [27].
There are several risk factors lead to emergence of CRC malignancies such as increased age, being a male as well as environmental and lifestyle related factors like smoking status, diet and obesity [10, 28]. Genetic mutations also play a pivotal role in CRC development which usually involves inactivation of tumor suppressor genes or activation of proto-oncogenes [29]. Moreover, mutations that reduce the ability of NER pathway to remove damaged DNA have also been shown to increase susceptibility to have tumors [30, 31].
The current study aimed to examine the possible association between ERCC2 genetic polymorphisms (rs1799793 and rs238406) and the risk of CRC emergence. rs1799793 is associated with lower DNA repair efficiency and one of the main causes of CRC carcinogenesis is genetic instability and lower DNA repairing capacity [32]. Although rs238406 results in silent polymorphism, it affects ERCC2 protein level indirectly [33]. As far as we know these two SNPs have never been investigated among Iraqi population with any other disease.
No association was observed between Asp312Asn and rs238406 genotypes, alleles or haplotypes with CRC susceptibility. In addition, the current research failed to find a relationship between ERCC2 variants and clinicopathological characteristics of CRC patients.
To our knowledge, only few research has found an association between rs238406 and increased susceptibility to CRC development. CC genotype was associated with a significantly elevated CRC susceptibility, with an odds ratio (95% CI) of 1.5 (1.1–2.0) [19]. However, a polish study revealed that AA genotype was associated with increased CRC risk. One the other hand, CC genotype was protective against CRC emergence [20]. Nevertheless, meta-analysis and bioinformatic analysis was published in 2019 concluded that rs238406 genotypes had lack of association with digestive system cancers, which is consistent with the present study findings [34]. The silent polymorphism of rs238406 does not lead to amino acid conversions. It could plausibly change the mRNA stability or disrupt protein synthesis in the 5′ proximal region of gene by altering a high-usage codon into a low-usage codon [35].
With regard to rs1799793, several studies assessed its relationship with CRC emergence with mixed results; Paszkowska-Szczur et al. concluded that AA genotype elevated the risk by approximately 3.5 times compared to wild type genotype carrier. When further Stratified analysis by gender and age was done both AA and AC genotypes increased the risk of CRC development in males and patients older than 50 years [20]. Additionally, a meta-analysis was conducted in 2022 to assess the impact of NER genes on CRC revealed that A allele carriers raise the risk of developing colorectal malignancy and further stratification analysis showed a consistent result in Caucasian population [36]. However, A Mexican study conducted on 108 patients and 119 controls revealed lack of association with CRC risk which is in agreement with the present research results [37]. Furthermore, one evidence of association was revealed in a meta-analysis investigated the role of Asp312Asn variant in CRC risk performed on 5740 CRC patients and 7135 healthy controls [38]. Moreover, there are other research with similar results [22, 23, 39].
As far as one can tell, there is only some haplotype information available for the assessment of the disease susceptibility between ERCC2 and CRC. In this study haplotype analysis was examined between the two SNPs however it did not reach the level of significance.
The conflict in findings between various research is explained by different ethnicities, relatively small sample size and a possible weak impact of these polymorphisms on CRC risk. Further studies are warranted with a larger sample size considering all other environmental risk factors and other genes in NER pathway.
CONCLUSIONS
The present study concluded lack of association between ERCC2 polymorphisms with risk of CRC development among a sample of Iraqi population. Nonetheless, the current study had a major limitation which is relatively small sample size. Therefore, extensive future investigation should be carried out as finding the role of potential genetic polymorphisms in CRC development will help with applying certain strategies to reveal individuals at risk among Iraqi populations.
ACKNOWLEDGEMENT
The authors would like to express appreciation to all subjects whose participation made this study possible. This research received no external funding.
AUTHOR CONTRIBUTIONS
The work was designed, supervised, and performed by RF, ESS, and AZA. The first draft of this manuscript was prepared by RF and ESS. RF analyzed the data and improved the overview of the manuscript. Collecting data was done by RF and AZA. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Jiang J, Zhang X, et al. Polymorphisms of DNA repair genes: Adprt, xrcc1, and xpd and cancer risk in genetic epidemiology. Methods Mol Biol. 2009;471:305-33.
- [2]Manuguerra M, Saletta F, et al. Xrcc3 and xpd/ercc2 single nucleotide polymorphisms and the risk of cancer: A huge review. Am J Epidemiol. 2006;164:297-302.
- [3]Benhamou S, Sarasin A. Ercc2/xpd gene polymorphisms and cancer risk. Mutagenesis. 2002;17:463-9.
- [4]Sameer AS, Nissar S. Xpd–the lynchpin of ner: Molecule, gene, polymorphisms, and role in colorectal carcinogenesis. Frontiers in Molecular Biosciences. 2018;5:23.
- [5]Itin PH, Sarasin A, et al. Trichothiodystrophy: Update on the sulfur-deficient brittle hair syndromes. J Am Acad Dermatol. 2001;44:891-920; quiz 1-4.
- [6]Hossain MS, Karuniawati H, et al. Colorectal cancer: A review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers (Basel). 2022;14.
- [7]Globocan. Colorectal cancer. International Agency for Research on Cancer; 2020.
- [8]Al Alwan NA. Cancer control and oncology care in iraq. J Contemp Med Sci. 2022;8:82-5.
- [9]Hussain AM, Lafta RK. Cancer trends in iraq 2000-2016. Oman Med J. 2021;36:e219.
- [10]Dhahir NK, Noaman, A. A. A comparative study of colorectal cancer based on patient’s age. Journal of the faculty of medicine Baghdad. 2021;63:70-3.
- [11]Dekker E, Tanis PJ, et al. Pure-amc. Lancet. 2019;394:1467-80.
- [12]Hanon BM, Al-Mohaimen Mohammad NA, et al. Cpg island methylator phenotype (cimp) correlation with clinical and morphological feature of colorectal cancer in iraq patients. Pan Arab Journal of Oncology. 2015;8.
- [13]Mahood WS, Nadir MI, et al. Methylation status of p16 gene in iraqi colorectal cancer patients. Iraqi journal of biotechnology. 2014;13:237-47.
- [14]Vuong LD, Nguyen HV, et al. Aberrant methylation of cdkn2a, rassf1a and wif1 in sporadic adenocarcinomatous colorectal cancer: Associations with clinicopathological features. J Adv Biotechnol Exp Ther. 2021; 4(3): 305-310.
- [15]Linh Dieu Vuong HHC, Quang Ngoc Nguyen. Study on relationship between genetic abnormalities and clinicopathological features in k hospital’s patients with colorectal cancer. J Adv Biotechnol Exp Ther. 2022;5:408-16.
- [16]Wu K-G, He X-F, et al. Association between the xpd/ercc2 lys751gln polymorphism and risk of cancer: Evidence from 224 case–control studies. Tumor Biology. 2014;35:11243-59.
- [17]Du H, Guo N, et al. The effect of xpd polymorphisms on digestive tract cancers risk: A meta-analysis. PLoS One. 2014;9:e96301.
- [18]Zhang S, Chen Z, et al. Genetic polymorphisms of DNA repair genes xpd, xpc, and xrcc4 in relation to colorectal cancer susceptibility. Chinese Journal of Clinical Oncology. 2017:365-70.
- [19]Salimzadeh H, Lindskog EB, et al. Association of DNA repair gene variants with colorectal cancer: Risk, toxicity, and survival. BMC cancer. 2020;20:1-10.
- [20]Paszkowska-Szczur K, Scott RJ, et al. Polymorphisms in nucleotide excision repair genes and susceptibility to colorectal cancer in the polish population. Mol Biol Rep. 2015;42:755-64.
- [21]Ho V, Peacock S, et al. Meat-derived carcinogens, genetic susceptibility and colorectal adenoma risk. Genes Nutr. 2014;9:430.
- [22]Chang W-S, Yueh T-C, et al. Contribution of DNA repair xeroderma pigmentosum group d genotypes to colorectal cancer risk in taiwan. Anticancer Research. 2016;36:1657-63.
- [23]Gil J, Ramsey D, et al. The c/a polymorphism in intron 11 of the xpc gene plays a crucial role in the modulation of an individual's susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012;39:527-34.
- [24]Alrubaie A, Alkhalidi N, et al. A clinical study of newly-diagnosed colorectal cancer over 2 years in a gastroenterology center in iraq. Journal of Coloproctology (Rio de Janeiro). 2019;39:217-22.
- [25]AJCC AJCoC. Colon and rectum. Ajcc cancer staging manual. 8th ed. New York: Springer; 2017.
- [26]Li Z ZZ, He Z, Tang W, Li T, Zeng Z, He L, Shi Y. A partition-ligation-combination-subdivision em algorithm for haplotype inference with multiallelic markers: Update of the shesis. 2009.
- [27]Mahmood AH, Zeiny SM, et al. Serological markers “cea test & sapril test” in iraqi patients with colon cancer. Journal of the Faculty of Medicine Baghdad. 2017;59:317-20.
- [28]Keum N, Giovannucci E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nature reviews Gastroenterology & hepatology. 2019;16:713-32.
- [29]Abbood IS, Aziz IH. Genetic variation in braf gene among iraqi colorectal cancer patients. Iraqi journal of biotechnology. 2018;17.
- [30]Toma M, Beluşică L, et al. Rating the environmental and genetic risk factors for colorectal cancer. Journal of medicine and life. 2012;5:152.
- [31]Al-Ward MA, Jawad MM. Study of multiple genotypes in the xpd gene for a/c lys751gln and lung cancer in samples from baghdad–iraq. Prof(Dr) RK Sharma. 2021;21:1727.
- [32]Ni M, Zhang W-z, et al. Association of ercc1 and ercc2 polymorphisms with colorectal cancer risk in a chinese population. Scientific reports. 2014;4:1-4.
- [33]Zhu ML, He J, et al. Potentially functional polymorphisms in the ercc2 gene and risk of esophageal squamous cell carcinoma in chinese populations. Sci Rep. 2014;4:6281.
- [34]Wu H, Li S, et al. Associations of mrna expression of DNA repair genes and genetic polymorphisms with cancer risk: A bioinformatics analysis and meta-analysis. J Cancer. 2019;10:3593-607.
- [35]Yin J, Wang C, et al. No evidence of association between the synonymous polymorphisms in xrcc1 and ercc2 and breast cancer susceptibility among nonsmoking chinese. Gene. 2012;503:118-22.
- [36]Yi C, Li T, et al. Polymorphisms of nucleotide excision repair genes associated with colorectal cancer risk: Meta-analysis and trial sequential analysis. Front Genet. 2022;13:1009938.
- [37]Gómez-Díaz B, M DLLA-M, et al. Analysis of ercc1 and ercc2 gene variants in osteosarcoma, colorectal and breast cancer. Oncol Lett. 2015;9:1657-61.
- [38]Ma X, Zhang B, et al. Genetic variants associated with colorectal cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut. 2014;63:326-36.
- [39]Zhang Y, Ding D, et al. Lack of association between xpd lys751gln and asp312asn polymorphisms and colorectal cancer risk: A meta-analysis of case–control studies. International journal of colorectal disease. 2011;26:1257-64.