Isolation and identification of Salmonella spp. and Escherichia coli from water used during live transportation of Pangasius catfish, Pangasianodon hypophthalmus
Abstract
Pangasius catfish (Pangasianodon hypophthalmus) is popular among fish farmers of Bangladesh due to its hardy characteristics, fast growth, and its ability to survive in high densities. Many consumers love to buy this fish, especially as live condition, due to its low market price and delicious fleshy meat. Bacterial outgrowth in transport water is frequent consequence including some enteric groups like Salmonella spp. and E. coli. The study attempted to know the occurrence of Salmonella spp. and E. coli in water used during live transportation of Pangasius catfish in Bangladesh. Water samples were collected from three different Pangasius catfish transportation channels at 2 h interval on-board transportation vehicle. The collected waters were then plated onto SS and EMB agar plates and 15, 20, and 17 suspected isolates were obtained from channel 1, 2 and 3, respectively. The isolates were confirmed through PCR techniques; Salmonella spp. was found only in channel 1 while E. coli were found in all 3 sampling channels under investigation. Among the suspected isolates, 13 isolates were positive for E. coli in channel 1, while 16 in both channel 2 and 3. Among the suspected isolates, 86.54% was E. coli positive, 1.92% was Salmonella positive, and 11.54% isolates were unidentified. The results indicated that the fishes were contaminated with Salmonella spp. and E. coli species either in the culture systems or during handling and live transportation.
INTRODUCTION
Pangasius catfish (Pangaisianodon hypophthalmus; Sauvage, 1878) is a freshwater benthopelagic fish of the Pangasiidae family and has been recognized as a commercial aquaculture species in many Asian countries including Bangladesh. Currently, the species contributes 18.37% of total aquaculture production of Bangladesh [1]. As Pangasius catfish is popular to consumers due to its low market value [2], it is transported in live condition from production sites to markets in several areas of Bangladesh [3]. Live transportation of Pangasius catfish includes harvesting it from the culture system, holding it in a confined plastic tank with water and carrying it to the desired retail market. Live transportation results in significant degradation of water quality parameters [4, 5], thus it may cause deteriorative changes in the fishes. Bacteria grow in the transport water can even make the water quality worse for fish during live transportation. Previous reports showed that the viable bacterial counts of the transport water increased significantly with the periods of live transportation of Pangasius catfish [3] and climbing perch, Anabas testudineus [6] in Bangladesh.
Bacteria of enteric origin (Enterobacteriaceae) are commonly disseminating from the gastrointestinal tract of humans and other animals are reported as common in aquatic environments [7, 8], which may normally present in different parts of the apparently healthy fishes from contaminated waters. Among the Enterobacteriaceae species, Salmonella, Escherichia coli, and Yarsenia enterocolitica are not typically found in water or aquatic goods [9,10]. Salmonella was not first identified in fish and is not a biological contaminant. It enters food through polluted water or inappropriate handling [11], and both Salmonella and E. coli are regarded as public health hazards because they can cause food poisoning. The location, cultivated species, breeding procedures, processing, and cultural practices are the key elements that affect the risk of microbial contamination in aquaculture products. Some of these possible microbial risks could be brought on by subpar hygiene standards, sewage, and livestock drainage. Leaching, for instance, introduces environmental toxins into river waters, where they end up in the fish and have detrimental impacts on this ecosystem [12]. In addition, massive use of fertilizer in the fish culture system and rearing fish with other types of animals, such as poultry, cattle and pigs are also responsible for contaminating the fish with Salmonella spp. [13-15].
The wide range of human diseases caused by Salmonella includes, enteric fever, bacteremia and gastroenteritis [16]. Salmonella is a second leading cause of foodborne illness worldwide [17]. The majority of human gastroenteritis by the Salmonella is caused through the ingestion of undercooked eggs, shellfish and fish [18]. Different studies have been reported that freshwater fishes are contaminated by Salmonella spp. from the area where they were reared [19]. Salmonella infections in freshwater fish are typically caused by faecal contamination of the water where the fish were caught [19]. High prevalence of Salmonella in catfish was reported [20]. The high temperature of the pond water, which enhances the organism’s growth rate, was accounted for the high prevalence rate [21]. The most prevalent coliform in the intestinal flora of warm-blooded animals, however, is E. coli, which is assumed to be mostly related to faecal contamination [22]. As a result, the potential that fish could serve as carriers of human pathogenic bacteria is receiving more attention [23-25], as a wide range of bacteria including Salmonella spp. and E. coli have been isolated from skin, digestive tracts, kidney and muscle of different fish species of both temperate and tropical waters [26, 27]. As common practice, underground water is used during live transportation of the fishes in Bangladesh [28] and Salmonella spp. was not identified in the skin and muscle of Pangasius catfish from several ponds in Bangladesh, while it was isolated from the same species after marketing. On the other hand, E. coli was found in the skin and muscle of Pangasius catfish before and after marketing [28]. Thus, it is not clear how the fishes are contaminated by these two pathogens of enteric origin. We also do not know whether the fishes are contaminated even during live transportation.
Based on the above background, the objective of this investigation was to assess the presence of Salmonella spp. and E. coli in water used during live transportation of the Pangasius catfish in Bangladesh. Investigations were done in three supply channels of Pangasius catfish and water samples were evaluated at 2 h intervals from the loading of the fishes.
MATERIALS AND METHODS
Descriptions of the studied supply channels of the Pangasius catfish
The study was conducted in three different Pangasius catfish supply channels of Bangladesh from July to December 2019. As Trishal Upazila of Mymensingh district is one of the major Pangasius catfish producing sites in Bangladesh, all the channels started from this Upazila. The supply channels were designated as “Channel 1” from Trishal, Mymensingh to Dhaka, “Channel 2” from Mymensingh to Faridpur, and “Channel 3” from Mymensingh to Sylhet (Figure 1).
Harvesting and preparation before live transportation
Pangasius catfish were harvested with the surrounding net and prepared for transportation at dusk. A very brief conditioning period was allowed prior to transportation. In most cases, transportation started at night and reached to the final destination/ unloading points (retail markets) at dawn. Around 40 kg fishes (20-22 fishes) were loaded in plastic made transportation tank having a capacity of 1000 liters. But the tanks were filled half with 500 liters of deep tube-well water. Approximately 40-42 transportation tanks were incorporated in a commercial vehicle (Truck). Exchange of water of transportation tank with the deep tube-well water once after 2-3 hours during live transportation of Pangasius catfish was commonly practiced in all the supply channels during transportation. At the retail market, Pangasius catfish were collected from the transportation tank and live fishes were separated from the dead ones.
Isolation of Salmonella spp. and E. coli
The isolation and identification of Salmonella spp. and E. coli was carried out based on culture on SS and EMB agar plates with slight modifications as described [29]. Plates with SS agar and EMB agar (HIMEDIA, India) were prepared and used for the isolation of enteric bacteria from water used during live transportation of Pangasius catfish. Water samples were collected from the Pangasius catfish transportation tank with previously sterilized plastic bottle from the start of transportation (loading) to the end (unloading) at every 2 h interval. After collection, water samples were spread on previously prepared SS and EMB agar plates immediately. The plates were brought to the Laboratory of Fisheries Microbiology, Department of Fisheries Technology, Bangladesh Agricultural University, Mymensingh, Bangladesh and incubated at 37 for 24 h. After incubation, colourless or translucent, black, pink-coloured colonies were observed on SS agar. Suspected colonies on EMB agar had black or dark centre with or without green metallic sheen. The resulted colonies from SS and EMB agar plates were subjected to subcultures again on SS and EMB agar plates in order to obtain isolated colonies for pure culture. The resulted isolated colonies were kept on previously prepared agar slant of plate count agar (HIMEDIA, India) for further analysis. The preserved colonies were then streaked on SS agar plate from the agar slant and the plates were incubated at 37 for 24 h. After incubation, the resulted colonies were used for identification of Salmonella spp. and E. coli.
Identification of Salmonella spp. and E. coli
Previously obtained isolates were incubated in nutrient broth for 24 h at 37 in order to increase the number of bacteria to extract DNA. For this purpose, a pure colony was transferred in a test tube containing 10 ml of previously prepared nutrient agar broth. The broth was then incubated at 37 for 24 h. After incubation, turbidity of the nutrient broth ensured the growth of bacteria. Then 1 ml of nutrient broth was taken in a sterilized eppendorf and centrifuged at 5,000 rpm for 3 min. The settlings on the eppendorf were mixed with 100 μl distilled water and again centrifuged at 5,000 rpm for 3 min. After that, the settlings were mixed with 100 μl distilled water and homogenized and subjected to boiling for 10 min followed by keeping in ice for cold shock. Again, centrifugation was done at 10,000 rpm for 10 min. Finally, the supernatant was collected and used as DNA template for PCR.
PCR amplification of extracted bacterial DNA
Two sets of pre-tested primers were used for identification of Salmonella spp. and E. coli (Table 1). For Salmonella spp., invA gene was targeted, and used forward primer, InvA F (ATCAGTACCAGTCGTCTTATCTTGAT) and reverse primer, InvA R (TCTGTTTACCGGGCATACCAT) for amplification [27]. While 16S rRNA gene was partially amplified using EC-1 (GACCTCGGTTTAGTTCACAGA) as forward primer and EC-2 (CACAGCTGACGCTGACCA) as reverse primer [30]. For PCR amplification, 12.5 μl of master mix, 8.5 μl of nuclease free DEPC treated dH2O, 1 μl forward primer, 1 μl reverse primer and 2 μl of extracted bacterial genomic DNA were taken into the PCR tubes. The tubes were placed into the thermal cycler (2720 Thermal Cycler, Applied Biosystems, Waltham, USA) immediately after adding the master mix with the DNA, and the cyclic program was resumed after the program was over. PCR products were visualization by agarose gel-electrophoresis.
Agarose gel electrophoresis of PCR product
For electrophoresis, 1.5% agarose (Sigma-Aldrich, USA) gel was used for electrophoresis of the PCR products. Gel casting tray was assembled with gel comb of appropriate teeth size and number. 1.5% agarose solution was prepared in TAE buffer by melting in a microwave oven. Melted agarose was poured onto the casting tray and allowed to solidify on the bench. The hardened gel in its tray was transferred to the electrophoresis tank containing sufficient TAE buffer to cover the gel. The comb was gently removed. Then, 5 μl of each PCR product was mixed with 1 μl loading buffer and the sample was loaded to the appropriate well of the gel. In addition, 5 μl DNA size marker was loaded in one well. The leads of the electrophoresis apparatus were connected to the power supply and the electrophoresis was run at 100V. When DNA migrated sufficiently as judged from the migration of bromophenicol blue of loading buffer, the power supply was disconnected. The gel was stained in ethidium bromide (0.5 μg/ml) for 10 min in a dark place. Then the gel was de-stained in distilled water for 10 min. De-stained gel was then placed on the UV transilluminator (Biometra, Germany) in the dark chamber of the image documentation system. The UV light of the system was switched on and the image was viewed on the monitor, focused, acquired, and saved in an USB flash drive.
RESULTS
Isolation of Salmonella spp. and E. coli
In order to detect Salmonella spp. and E. coli, water used in live transportation of Pangasius catfish was collected from three marketing channels. The collected water samples were then inoculated onto SS and EMB agar plates and well-separated colonies were obtained, regarded as isolates. A total of 52 isolates were collected depending on the characteristics of the colony on SS and EMB agar plates.
Identification of Salmonella spp. and E. coli
The DNA of all the isolates were extracted and subjected to PCR according to the protocol described in the methodology section. Predefined invA genes of Salmonella spp. and parts of 16S rRNA genes of E. coli were targeted using two different sets of primers namely, InvA (211 bp) and ECO (585 bp) for Salmonella spp. and E. coli, respectively (Table 1). Salmonella spp. positive was confirmed considering 211 bp bands on gel after electrophoresis. For E. coli, bands of 585 bp on gel were considered positive (Figure 2 and 3).
Table 1. PCR protocol used for the confirmation of Salmonella spp. and E. coli.
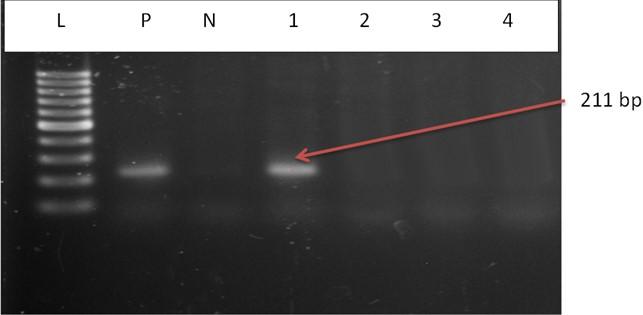
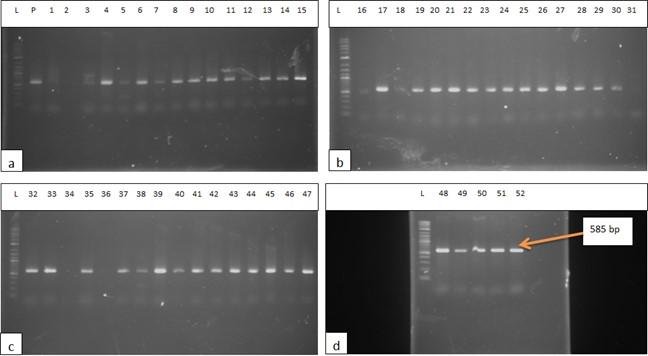
Incidence of Salmonella spp. and E. coli in water used during live transportation of Pangasius catfish
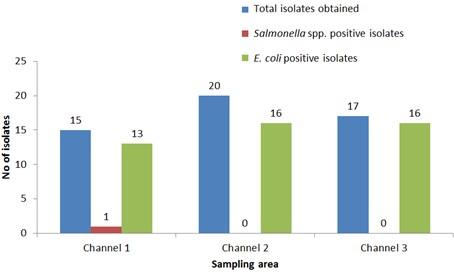
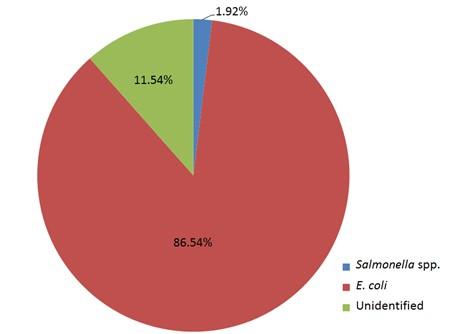
Out of 15 isolates from sampling Channel 1, only 1 isolate (6.67%) was positive for Salmonella spp., while 13 isolates (86.66%) were positive for E. coli and 1 isolate (6.67%) remained unidentified. In Channel 2, 16 E. coli positive isolates were identified which accounts for 80% of the total collected isolates and 4 isolates (20%) remained unidentified. In sampling Channel 3 from 17 isolates, 16 isolates (94.12%) were identified as positive for E. coli and 1 isolate (5.88%) remained unidentified. On the other hand, 45 isolates (86.54%) were identified as positive for E. coli and the remaining 6 isolates (11.54%) were not confirmed as Salmonella spp. or E. coli, gave no band on electrophoresis (Table 2, and Figure 4). Although, E. coli positive isolates were found in all the three sampling channels, only one Salmonella spp. positive isolate was confirmed obtained from the sampling Channel 1 (Table 2 and Figure 5).
Number of isolates obtained from water used during live transportation of Pangasius catfish (Pangasianodon hypophthalmus) in Bangladesh. The water samples were collected from 3 different supply channels.
Table 2. Positive isolates for Salmonella spp. and E. coli.
DISCUSSION
A previous study showed the presence of both Salmonella spp. and E. coli in Pangasius catfish collected from several retail markets regardless of their presence or absence in the same specimen before marketing. So, the experiment was designed to identifying the presence of Salmonella spp. and E. coli in water which was used in live transportation of Pangasius catfish from farms to retail markets. In order to achieve the objectives of the study, water samples were collected from the plastic barrels where Pangasius catfish were kept alive during transportation from farm to the retail markets at every 2 h interval from 3 different Pangasius catfish marketing channels of Bangladesh. We assumed that these two pathogens would be detected from the water subsamples collected from the barrels used in live transportation. The detection of Salmonella spp. and E. coli from the water used in live transportation of Pangasius catfish is of great concern due to its potential to cause enteric disease [31]. The water samples were taken aseptically from the plastic barrels where Pangasius catfish were carried during transportation. After collection, water samples were then placed on SS and EMB agar plates at the start of transportation to the end at 2 h interval. Upon completion of sampling, plates were brought to the laboratory and incubated at 37 for 24 h. After growth of microorganisms on SS and EMB agar plates, suspected Salmonella spp. and E. coli colonies were isolated according to a protocol with slight modifications as described [29]. The collected isolates of Salmonella spp. and E. coli from subsamples water were identified through PCR using two primers which are very fast, unique and sensitive in identifying the target gene by PCR technique [32].
Highest incidence of E. coli was found in sampling Channel 3 and the lowest was in sampling Channel 2. Incidence of Salmonella spp. was seen only in the sampling Channel 1. Among the collected isolates regardless of the sampling channels, 86.54% were identified as positive for E. coli whereas only 1.92% Salmonella spp. positive isolates were identified. On the other hand, 11.54% isolates remained unidentified. In comparison to Salmonella spp., percentages of E. coli positive isolates were always higher in the studied channels. Fish act as carrier of microorganisms as it is continuously exposed to the microorganisms in aquatic environment and the microorganism in fish reflects the conditions of the environment. At the time of harvesting, a wide variety of microorganisms contain in the body of fresh fish which are known as the microflora of that fish [33]. Fish or processed fish may be contaminated with different types of bacteria, such as Salmonella, coliform, faecal coliform, Streptococcus, Staphylococcus aureus and these are responsible for different types of foodborne disease [34]. In our experiment, Salmonella spp. was found only in sampling Channel 1, while E. coli was identified from all the sampling channels. Identification of these enteric bacteria indicated that the water was polluted with faecal matters. Bird droppings, human faeces and other animal faeces falling into aquatic environment are contaminated with enteric bacteria. Deep tube well water was used during live transportation of Pangasius catfish. In addition, stated that ground water was free from any sorts of coliform organisms, and they isolated E. coli from the pond sediments and stated the sources as pigeon [35]. Moreover, a study found E. coli in all the tube well water samples in their study while Salmonella spp. was absent in all tube well water samples [26]. On the other hand, low concentration of faecal coliform bacteria in tube well water of Bangladesh [36]. Groundwater can also be contaminated by a wide range of pathogens [15, 37-38] and found Salmonella spp., and E. coli in ground water sample. Identified enteric bacteria would be the reflection of the ponds from where the Pangasius catfish were harvested. The presence of enteric bacteria in water, sediment and fish reflects the water used in transportation are transmitted by fish. Moreover, Unhygienic handling of Pangasius catfish during transportation may contaminate the fish with Salmonella spp. and E. coli. [28]. It also stated that rough handling and lack of proper sanitation during transportation causes Pangasius and other fishes more contaminated collected from retail market compared to pond samples of the same species.
CONCLUSIONS
Bacterial hazards are closely related to food safety. The prevalence of Salmonella spp. was lower in Pangasius catfish transportation water in comparison to E. coli. The presence of Salmonella spp., and E. coli in water used during live transportation of Pangasius catfish indicated that the fishes were harvested from water which was polluted with faeces. Water used in the transportation would be another source of enteric bacteria. Personnel involved in handling of fish during live transportation would be a potential source of enteric bacteria. These enteric microorganisms could cause foodborne illness among the consumers which ultimately affects the profit margin of farmers. So, it is necessary to maintain proper sanitation and handling during live transportation of Pangasius catfish to ensure food safety.
ACKNOWLEDGEMENTS
The authors are grateful to the Department of Fisheries Technology, Bangladesh Agricultural University, for the laboratory and technical support and to the Bangladesh Agricultural University Research System (BAURES) for the research grants.
AUTHOR CONTRIBUTIONS
The work was designed and supervised by MNH and MNU. The research work was performed by ANMRKB and MMH. The first draft of this manuscript was prepared by ANMRKB. MMH analyzed the data and improved the overview of the manuscript. MNH critically revised, improved and approved the final version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]DoF. Yearbook of Fish Stat Bangladesh, 2017-18 Fish resour surv syst (FRSS), Dep Fish Bangladesh Minist Fish. 2018;35:129.
- [2]Belton B, Karim M, Thilsted S, Murshed-E-Jahan K, Collis W, Phillips M. Aquaculture and fish consumption in Bangladesh Project Leader. 2011.
- [3]Bhuiyan ANMRK, Hossain MM, Uddin MN, Hossain MA, Hossain MI, Haider MN. Changes in viable bacterial counts and physicochemical parameters of water used during live transportation of Pangasius catfish (Pangasianodon hypophthalmus) in Bangladesh. J Adv Vet Anim Res. 2022;9:66–77.
- [4]Berka R. The transport of live fish. A review. EIFAC Technical Paper 1986; 48, 52 pp.
- [5]McFarland WNTK, Norris S. The control of pH by buffers in fish transport.” California Fish and Game. 1958;4: 291-310.
- [6]Hossain MM, Bhuiyan ANMRK, Hossain MA, Uddin MN, Hossain MI, Haider MN. Follow up of bacterial and physicochemical quality of water during live transportation of climbing perch (Anabas testudineus) in Bangladesh. J Adv Biotechnol Exp Ther. 2021;4:149–60.
- [7]Newaj-Fyzul A, Mutani A, Ramsubhag A, Adesiyun A. Prevalence of bacterial pathogens and their anti-microbial resistance in tilapia and their pond water in Trinidad. Zoonoses Public Health. 2008;55:206–13.
- [8]Wogu M, Maduakor C. Evaluation of microbial spoilage of some aquacultured fresh fish in benin city Nigeria. Ethiop J Environ Stud Manag. 2011;3.
- [9]Aziz H, Daphn A. Bacteriological studies of faecal and water samples from different sources with special reference to some Gram-negative bacteria. Benha Vet. Med. J.,2005:248-61.
- [10]FAO. Assessment and management of seafood safety and quality. FAO fisheries technical paper 2003:444.
- [11]Pao C, Molla B, Kleer J, Reine A. Hygienic control of fish processing plant. Wochenschr. 2008;121(4):89-93.
- [12]Traore O, Nyholm O, Siitonen A, Bonkoungou IJO, Traore AS, Barro N. Prevalenceand diversity of Salmonella enterica in water, fish and lettuce in Ouagadougou, Burkina Faso. BMC Microbiol 2015;15:1–7.
- [13]Ampofo JA, Clerk GC. Diversity of bacteria contaminants in tissues of fish cultured in organic waste-fertilized ponds: Health Implications~!2009-10-15~!2010-02-16~!2010-06-17~! Open Fish Sci J. 2010;3:142–6.
- [14]Esposto EM, Silva WCP, Reis CMF, Reis EMF, Ribeiro R V., Rodrigues DP. Enteropatogenos bacterianos em peixes criados em uma estacao de reciclagem de nutrientes e no ecossistema relacionado. Pesqui Veterinária Bras. 2007;27:144–8.
- [15]Li K, Petersen G, Barco L, Hvidtfeldt K, Liu L, Dalsgaard A. Salmonella weltevreden in integrated and non-integrated tilapia aquaculture systems in Guangdong, China. Food Microbiol. 2017;65:19–24.
- [16]Nwiyi P, Onyeabor A. Occurrence of Salmonella spp from fresh fish (Tilapia Nilotica Linn) using improved isolation methods. Online J Anim Feed Res. 2012;2:475–8.
- [17]Wong MH, Chen S. First detection of oqAB in Salmonella spp. isolated from food. Antimicrobial agents and chemotherapy. 2013;;57:658-60.
- [18]Awuor WS, Miruka OD, Eliud WN. Characterisation of Salmonella isolated from nile tilapia (Oreochromis niloticus) along lake victoria beaches in western Kenya. Inter J Biol Med Sciences. 2011;1:51-6.
- [19]Mhango M, Mpuchane SF, Mpuchane BA. Incidence of indicator organisms, opportunistic and pathogenic bacteria in fish. African Journal of Food, Agriculture, Nutrition and Development. 2010;10:10.
- [20]Wyatt LE, Nickelson R, Vanderzant C. Occurrence and control of Salmonella in freshwater catfish. Journal of Food Science. 1979;4:67-73.
- [21]Wyatt LE, Nickelson R, Vanderzant CA. Edwardsiella tarda in freshwater catfish and their environment. Applied and Environmental Microbiology. 1979;38:710-4.
- [22]Rompre A, Servais P, Baudart J, Roubin MR, Laurent P. Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. Journal of microbiological methods. 2002;49:31-54.
- [23]Buras N, Duek L, Niv S, Hepher B, Sandbank E. Microbiological aspects of fish grown in treated wastewater. Water Res 1987;21:1–10.
- [24]Hejkal TW, Gerba CP, Henderson S, Freeze M. Bacteriological, virological and chemical evaluation of a wastewater-aquaculture system. Water Res 1983;17:1749–55.
- [25]Hejkal TW, Gerba CP, Henderson S, Freeze M. Bacteriological, virological and chemical evaluation of a wastewater-aquaculture system. Water Res 1983;17:1749–55.
- [26]Sarker S, Mahmud S, Sultana R, Biswas R, Sarkar PP, Munayem MA. Quality assessment of surface and drinking water of nakla paurosova, Sherpur, Bangladesh. Adv Microbiol 2019;09:703–27.
- [27]Ogunremi D, Davis S, Dupras AA, Marquez IG, Omidi K, Pope L. Evaluation of a multiplex pcr assay for the identification of Salmonella serovars enteritidis and typhimurium using retail and abattoir samples. J Food Prot 2017;80:295–301.
- [28]Hasan GMMA, Hossain MS, Parveen S, Juliana FM. Microbiological and chemical quality assessment of six fish species of Bangladesh during freeze storage. Int J Res Appl Sci Eng Technol 2016;4:572–8.
- [29]Haider MN, Faridullah M, Kamal M, Islam MN, Khan MN. A bacteriological assessment for Salmonella and Escherichia coli in some selected freshwater prawn (Macrobrachium rosenbergii) farms and depots. Journal of Marine Bioscience and Biotechnology. 2007;2:40-7.
- [30]Hossain M, Siddique M, Hossain F, Zinnah M, Hossain M, Alam M. Isolation, identification, toxin profile and antibiogram of Escherichia coli isolated from broilers and layers in Mymensingh district of Bangladesh. Bangladesh J Vet Med 1970;6:1–5.
- [31]Oh JY, Kang MS, Kim JM, An BK, Song EA, Kim JY. Characterization of Escherichia coli isolates from laying hens with colibacillosis on 2 commercial egg-producing farms in Korea. Poult Sci 2011;90:1948–54.
- [32]Yanestria SM, Rahmaniar RP, Wibisono FJ, Effendi MH. Detection of invA gene of Salmonella from milkfish (Chanos chanos) at Sidoarjo wet fish market, Indonesia, using polymerase chain reaction technique. Vet World 2019;12:170–5.
- [33]Trust TJ, Sparrow RA. The bacterial flora in the alimentary tract of freshwater salmonid fishes. Canadian Journal of Microbiology. 1974;20:1219-28.
- [34][34] Mobin SM, Chowdhury MB, Islam MS, Uddin MN. Status of bacterial flora in the intestine of two freshwater fish. Bangladesh J. Life Sci. 2001;13:149-55.
- [35]Al-Harbi AH, Uddin MN. Seasonal variation in the intestinal bacterial flora of hybrid tilapia (Oreochromis niloticus x Oreochromis aureus) cultured in earthen ponds in Saudi Arabia. Aquaculture 2004;229:37–44.
- [36]Islam MS, Siddika A, Khan MNH, Goldar MM, Sadique MA, Kabir ANMH. Microbiological analysis of tube-well water in a rural area of Bangladesh. Appl Environ Microbiol 2001;67:3328–30.
- [37]John DE, Rose JB. Review of factors affecting microbial survival in groundwater. Environ Sci Technol 2005;39:7345–56.
- [38]Suthar S, Chhimpa V, Singh S. Bacterial contamination in drinking water: a case study in rural areas of northern Rajasthan, India. Environ Monit Assess 2009;159:43–50.