Molecular characterization and antibacterial activities of mangrove endophytic fungi from coastal Kenya
Abstract
The advent of resistant pathogenic bacteria and fungi across the globe is threatening the efficacy of antibiotic drugs. Thus, microbial infections are becoming a threat to life. Endophytic fungi remain a viable source of secondary metabolites with unique spectra of biological activities. This study isolated and characterized endophytic fungi from selected mangrove species of coastal Kenya and further ascertained their activities. A total of 18 fungal endophytes selected from mangrove species were investigated for antimicrobial activity against gram-positive Staphylococcus aureus and gram-negative Escherichia coli. Potato dextrose agar and potato dextrose broth were used for isolation, purification, and fermentation at 28oC for 7–15 days. Extraction of fungal metabolites was achieved using ethyl acetate (1:1 v/v) and ethyl acetate in 10% methanol (9:1 v/v). Solvents were recovered in a fume hood and extracts were dissolved in 1 ml of dimethyl sulfoxide. Molecular characterization completely identified 9 species, namely: Aspergillus flavus, Aspergillus niger, Aspergillus tubingensis, Aspergillus oryzae, Rhizophora nomius, Aspergillus awamori, Aspergillus aculeatus, Aspergillus bravionivious, and Aspergillus welwitchiae. The minimum inhibitory concentration of ethyl acetate crude extracts of the most active fungal isolate, A. flavus (MT447532.1), was 0.91 ± 0.05 mg/ml and 0.82 ± 0.052 mg/ml against S. aureus and E. coli, respectively. Results showed that some crude extracts of mangrove fungal endophytes from coastal Kenya are effective against bacteria, hence a promising source of novel organic natural metabolites with a possible wide range of biological activities.
INTRODUCTION
There has been a rapid increase in multidrug-resistant (MDR) bacteria against antibiotics. This being a worldwide phenomenon, new resistance mechanisms are emerging and a rise in MDR is one of the reasons for the failure of treatment and increased deaths [1]. The emergence and spread of drug-resistant pathogens continue to threaten the treatment of common infections. The World Health Organization’s (WHO) reserves of novel antimicrobials are being depleted, making access to quality medicines a challenge [2]. To date, pathogenic strains of bacteria that were previously treatable through antibiotic therapy are resistant to every available antibiotic, while many other serious and pathogenic strains are resistant to all but one drug, thereby placing them on the edge of being deadly pathogens [3]. Staphylococcus aureus is an adaptable pathogen found in both animals and human beings and is responsible for increasing infections, starting with minor skin infections, and progressing to more serious and deadly ones such as meningitis and pneumonia. This pathogen is an important agent of food poisoning globally and is transmitted to humans through contaminated and unprocessed milk and milk products. It is resistant to penicillin, methicillin, and vancomycin [4, 5]. Escherichia coli is a rodlike gram-negative bacterium commonly occupying the lower intestine of mammals and whose transmission is mostly by fecal-oral-rout [6]. Most E. coli are innocuous normal flora in the gut that are beneficial to their host through the production of Vitamin K2 and B-Complex. These vitamins aid in preventing the establishment of bacteria within the intestinal walls. However, others, such as serotype O157:H7, cause food poisoning in humans, hemolytic uremic syndrome (HUS), and death in the elderly, the very young, and immunocompromised individuals. Antibiotic resistance is on the rise because of antibiotic overuse in humans and the rate of adaptive mutations in E. coli, an important factor for bacterial antibiotic resistance [6].
Natural products have been the utmost source of all medicines for most of human history [7]. Endophytic fungi inhabit an unmatched environmental position, being thought to impact the ecology, distribution of plants, physiology, and biochemistry [7]. Fungal endophytes have also been known as possible sources of innovative naturally occurring compounds applicable in agricultural, industrial, and pharmaceutical applications, especially those secondary metabolites produced by fungal endophytes inhabiting medicinal plants [8]. Mangrove endophytic fungi have become an important natural source for the discovery of novel biologically active compounds [9]. Mangroves are uniquely adapted to harsh environmental conditions like high salinity, anaerobic soils, and the rising sea levels [10]. They function as primary producers in the estuarine food chain, contributing to an ecological, economic, scientific, and cultural resource [10]. Most mangrove genera and families are not closely related but have common adaptive features. These include developed morphological, physiological, biological, and ecological adaptations to extreme environmental conditions such as changes in sea level, high salinity, high temperatures, and anaerobic soils [11]. This is made possible by the presence of such features including pneumatophores, stilt roots, salt-secreting leaves, and viviparous water-dispersed propagules [10]. In Kenya, there are approximately 54,000 ha of mangrove forests, most of which are found in north coast in the counties of Tana River and Lamu [12]. Endophytic fungi inhabit the internal tissues of host plants for all or part of their life cycle. Almost every plant species harbors non-mycorrhizal fungal endophytes [8]. Among the diverse species of mangroves, only 9 are reported to occur along the Kenya coast, where their ecological habitats are systematically being destroyed through human activities [13], thus threatening them with imminent extinction. The diversity of mangrove fungal endophytes of Rhizophora mucronata, Avicenia marina, Ceriops tagal, and Sonneratia alba were investigated based on cultural and morphological characteristics [14]. A total of 18 fungal isolates based on the cultural and morphological characterization have been reported [14].
The present study focused on molecular characterization of the mangrove endophytic fungi and their antibacterial potential of crude extracts from selected mangrove species.
MATERIALS AND METHODS
Description of the study area
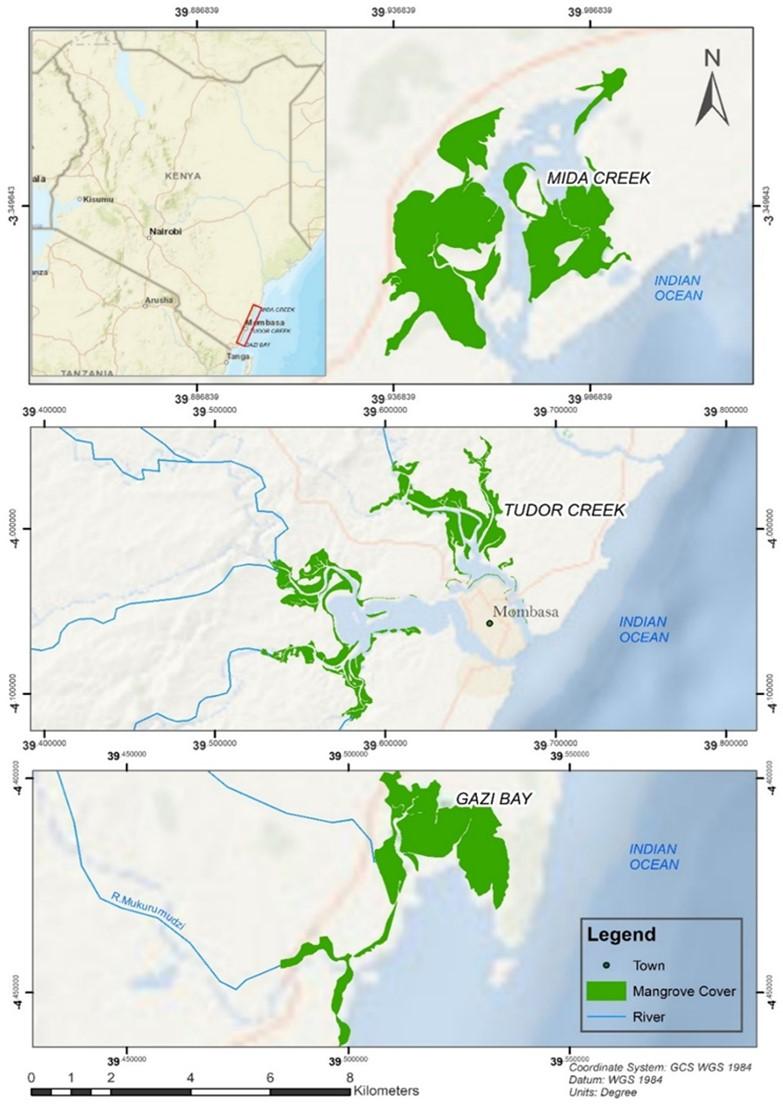
The study was conducted along the Kenya coast in three different mangrove forests (Figure 1). Gazi Bay is located on the south coast of Kenya, at 4°25’S and 39°50’E in Kwale County, about 50 km south of Mombasa. This bay is made up of a shallow channel measuring about 4 km long, bordered by mangrove forests dominated by the species Sonneratia alba and Rhizophora mucronata [15]. Tudor Creek is situated at 42’S, 3940’E on northwest of Mombasa Island and extends between 10–15 km inland. Two seasonal rivers, Kombeni and Tsalu, drain into the creek over 45,000 and 10,000 ha, respectively [16]. A mangrove forest within the creek extends over an area of 1, 1.64 ha, primarily made up of Sonneratia alba, Avicenia marina, and Rhizophora mucronata. The forest has different mangrove species zonation along the tidal gradient and is covered by deposits that are mostly made up of mud and sand. Parts of the forest are made up of mud and sand [17]. Mida Creek (03°21’S, 39°59’E) is situated in Kilifi County approximately 25 km south of Malindi town and about 88 km north of Mombasa City [16]. The mangrove forest is the most prominent habitat in Mida Creek and occupies approximately 1,746 ha. The forest supports 7 out of the 9 mangrove species found in Kenya [13].
Isolation of mangrove endophytic fungi
Stem twigs from selected mangrove species of Sonneratia alba, Ceriops tagal, Rhizophora mucronata, and Avicenia marina were collected and transported in airtight bags to the laboratory and stored. Surface sterilization was achieved by washing them in running tap water to remove debris. They were dipped in 70% for 3 min and then in 4% sodium hypochlorite for 3 min, followed by rinsing with sterile water for 2 min and drying under sterile conditions [14]. The outer tissues were removed using a laboratory knife after drying the plant tissues under sterile laminar air flow and passing through a flame. Internal tissues were split into smaller pieces of 0.5 to 1 cm under sterile conditions and plated on Potato Dextrose Agar (PDA) in order to obtain a wide range of fungal isolates. The plates were then incubated for 7-10 days at 28oC. Fungal growth was observed and monitored for the 10 days. Purity plating was done to get pure colonies, and it was done in triplicates for molecular characterization [14].
Molecular identification of the mangrove fungal endophytes
Mangrove fungal endophytes were selected for molecular characterization based on their growth characteristics such as color, colony form, mycelium, and colony margin. A total of 18 mangrove endophytic fungal isolates from the selected mangrove species of Sonneratia alba, Ceriops tagal, Rhizophora mucronata, and Avicenia marina were chosen for molecular identification. Total genomic deoxyribonucleic acid (DNA) extraction of selected mangrove fungal endophytes was done using the Quick-DNATM Fungal/Bacterial Mini Preparation Kit by Zymo Research, Catalog No. D6005 [18]. Prior to that, selected mangrove fungal endophytes were cultured in PDA at 28oC for 5 days. To remove any traces of DNA, all surfaces were cleaned with 3.85 % sodium hypochlorite, followed by 70% ethanol. The extraction buffers were carefully prepared using deionized water and later sterilized by autoclaving at 121oC for 15 minutes. All reusable apparatus was first UV-sterilized for 1 hour before being cleaned in between by soaking in 3.85% sodium hypochlorite for 30 minutes and thoroughly rinsing with sterilized distilled water. Beta-mercaptoethanol was added to the Genomic Lysis Buffer at a final concentration of 0.5% (v/v) to improve performance [18].
The target regions were amplified using universal primers for fungal DNA; internal transcribed spacers, ITS1 F-(5″-CTTGGTCATTTAGAGGAAGTAA-3″) and ITS4 (5″-TCCTCCGCTTATTGATATGC-3″). Agarose gel was prepared according to standard procedures and then mounted on the gel electrophoresis tank. 3 µl of the sample was mixed with a drop of loading dye and loaded into the wells, and electric current was run at 70 V for 50 minutes. The gel was then observed under a UV-Transilluminator, and gel images were captured. The concentration and purity of the DNA were confirmed using spectrophotometry (Eppendorf Bio spectrometer, software 4.3.5.0). From the agarose gel electrophoresis, DNA bands of sufficient thickness were observed, a preliminary indicator of a successful DNA extraction. All working areas were cleaned with 3.85% sodium hypochlorite to remove any traces of DNA, followed by 70% methanol for quality control. The polymerase chain reaction (PCR) amplicons were sequenced at Inqaba Boitech (South Africa). The DNA sequences obtained from ITS amplification were submitted to GenBank, and accession numbers were assigned to each fungus. DNA sequences were edited using UGENE version 36.0 and molecular evolutionary genetics analysis software for microorganisms (MEGA X).
Fermentation and extraction of fungal metabolites
All endophytic cultures were cultivated in 250 ml universal bottles containing 30 ml of Potato Dextrose Broth (PDB) and incubated at 28oC for about 3 weeks under static conditions. The broth culture was then filtered to separate the mycelia from the filtrate. The cell-free fermentation broths were extracted using two methods. In one method, the broth was homogenized in a 10% methanol/water mixture before being extracted in analytical grade (98% purity) ethyl acetate. The other method involved homogenization of the fungal broth and extraction with ethyl acetate. In each case, the fungal mycelia were separated from the filtrate by filtering them through four layers of muslin clothing. The cell-free fermentation broths were extracted three times with equal volumes of ethyl acetate. The mixture was left to stand for two hours and filtered to remove the mycelium residues. The filtrate was collected in a 50 ml centrifuge tube and centrifuged at 8,000 revolutions per minute (rpm) for 10 minutes to separate the organic phase and the broth phase [14]. The organic layer was removed by decantation and transferred to new tubes. An equal volume of ethyl acetate was added to the broth phase and extracted three more times. The resulting extracts were evaporated to dryness in a fume hood and gave a solid and oily residue. Extracts were dissolved in dimethyl sulfide oxide (DMSO) and used for antimicrobial assays. The DMSO extracts were kept at -20 °C for future research [10].
In vitro antibacterial susceptibility test
Ethyl acetate and methanolic crude extracts of the mangrove fungal endophytes dried in the fume hood were tested against S. aureus, American Type Culture Collection (ATCC 25922) and E. coli (ATCC 25922) obtained from Technical University of Mombasa, (TUM) Biology laboratory. Antimicrobial activities of the mangrove endophytic fungal crude extracts were determined by agar disk diffusion method. The test organisms were prepared in Mueller Hinton Agar Nutrient broth and incubated at 37 ºC for 24 hours and 3 days at 28 ± 1ºC, respectively [10]. Mueller Hinton Agar was prepared according to the manufacturer’s instructions. 32g of the Mueller Hinton agar was weighed and dissolved in 900 ml of distilled water. The mixture was heated to completely dissolve the medium, autoclaved at 121°C for 15 minutes and allowed to cool for 30 minutes. The media was poured on sterile petri dishes to 24 mm in a laminar flow hood to avoid any contamination and allowed to cool for a complete setting for 30 minutes. The plates were left ajar to avoid moisture build-up on the lid that could cause contamination. The plates were then appropriately labelled. After incubation, bacterial suspensions were spread on the surface of the solid agar medium (MHA) using sterile inoculating loop and left to dry for 5 minutes before being used for the antimicrobial assay [10]. The discs were placed on the dried solid agar medium using a sterile pair of forceps and firmly pressed on the surface to prevent falling off during incubation. Discs were evenly placed on the surface of the medium ensuring that a distance of 12-15 mm away was maintained from the edge of the plate and away from each other. Each petri plate contained one test sample, one positive control Amoxicillin and one negative control (DMSO). Lids were replaced, sealed with Parafilm and plates were incubated at 37ºC for 24 hours and activity was expressed as the diameter of the clear zone of inhibition (mm) around agar plugs indicating antimicrobial activity of the crude mangrove endophytic fungal extracts [19, 20].
Minimum inhibition concentrations of Aspergillus flavus extracts
Minimum inhibition concentrations (MICs) of the ethyl acetate crude extracts of Aspergillus flavus which displayed the strongest antimicrobial activity against S. aureus (ATCC 25922) and E. coli (ATCC 25922) were determined using micro-dilution method in a 96-well plate. The crude extract of the fungal isolate A. flavus was dissolved in DMSO and working concentrations made in 2-fold serial dilutions where the highest concentration was 1 mg/ml. The bacterial test organisms were grown on Müeller Hinton broth to 0.5 Macfarland standard. Test antibiotic solution was prepared as 1 g/ml then diluted in Müeller Hinton broth to 2-fold the top concentration desired in the test. 100 µL of Müeller Hinton broth was dispensed into all wells of the microtiter plate. 100 µL of 2-fold antibiotic solution was pipetted into the wells of column 1. Using the pipette, the antibiotic was mixed by sucking up and down 5-8 times. 100 µL of the mixture was withdrawn from column 1 and added to column 2 and then to column 3. The procedure was repeated up to column 10. 100 µL was discarded from column 10. 5 µL of bacteria was poured into wells from columns 1 to 11. No bacteria were added into column 12 serving as the blank. Plates were incubated at 37 °C for 18 hours. The reading of results was done in dual-mode microplate reader with monochromator-based optics for absorbance and sensitive top and bottom reading fluorescence applications (INFINITE M NANO) Pro200 Series, Austria at 600 nm. Experiments were performed in triplicate [21].
Data analysis
Phylogenetic analysis
The DNA sequences with assigned accession numbers and sequence matches from National Centre for Biological Information (NCBI) Basic Local Alignment Search Tool (BLAST) were retrieved and combined into a single text file in fasta format. Using UGENE, NCBI BLAST, the data base was queried using default settings and percentage identity threshold set at about 97% [22]. The sequences were aligned in Multi Sequence Alignment (MUSCLE) software found in MEGA X and manually trimmed before employing homology searches using BLASTn program against Gen Bank data base in NCBI [22]. The evolutionary history was inferred by using the Maximum Likelihood method and kimura 2-parameter model [23]. Initial trees (s) for the heuristic search were obtained automatically by applying Neighbour-Join and Bio NJ logarithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior long likelihood value. The analysis involved 19 nucleotide sequences and there was a total of 747 positions in the final dataset. Evolutionary analysis was conducted in MEGA X [24]. Data sequences for Fusarium oxysporum, a closely related species was retrieved from the database. A phylogenetic tree was constructed using the data sequences and reference to maximum likelihood method thereby analyzing the phylogenetic affiliations of the mangrove fungal endophytes [25]. Construction of the phylogenetic tree in MEGA X used defaulting parameters and bootstrap standards derived after 1000 replications.
Antimicrobial data analysis
A descriptive analysis of antibacterial activities and of ethyl acetate and methanolic extracts of mangrove endophytic fungi was performed and reported as mean Standard Deviation (SD) using Microsoft Excel. The MICs of ethyl acetate crude extracts against Staphylococcus aureus and Escherichia coli were determined and reported as mean Standard Error (SE). One-way ANOVA was used to compare the zone of inhibition of the fungal extracts against the positive and negative controls using STATISTICA statistical software version 7. The homoscedasticity of variances to meet the requirements of the analysis of variance (ANOVA) test was confirmed using Levene’s test at p > 0.05 and significance difference was assigned at p < 0.05.
RESULTS
Molecular identification of mangrove fungal endophytes
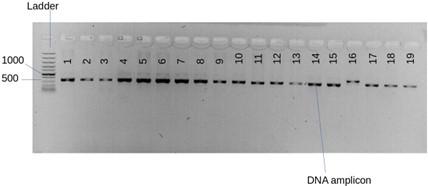
A total of 50 fungal isolates were originally obtained from twig cuttings of selected mangroves [14]. Bands of sufficient thickness were observed from the agarose gel electrophoresis and PCR amplicons (Figure 2) from PCR. The average range of DNA concentration was 244.8 μg ml-1 which was within the normal range at between 244.8 and 549.9 μg ml-1. The absorbance of DNA at A260 /A280 ranged at between 1.94 and 2.08. BLAST analysis gave closely related species (Table 1). 7 out 9 mangrove fungal isolates identified had 100 % similarity with the GenBank species. The 9 fungal isolates identified in this study were grouped into 3 clusters belonging to the phylum Ascomycota and genus Aspergillus.
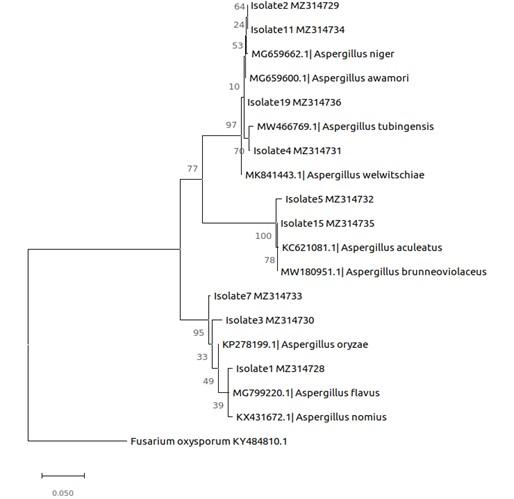
Results of phylogenetic analysis (Figure 3) showed that first cluster comprised of 3 isolates namely, AmT2, SaT1 and SaT3 grouped under Aspergillus with a 100 % bootstrap support representing Aspergillus oryzae (KP278199.1), A. flavus (MG799220.1) and A. nomius (KX431672.1) (Figure 3). The second cluster showed 2 isolates, CtT1 and AmM2 under the genus Aspergillus with 98.81 % and 98.93 % bootstrap support, respectively representing A. aculeatus (KC621081.1) and A. brunneoviolaceus (MW180951.1). The third cluster showed 4 isolates namely, SaT2, AmG1, SaT4 and RmM4 with a 100 % bootstrap support. The cluster was made of A. niger (MG659662.1), A. awamori (MG659600.1), A. tubingensis (MW466769.1) and A. welwitchiae (MK841443.1).
Phylogenetic analysis resulted in the tree with the highest log likelihood (-2246.83) (Figure 3). The percentage of trees in which the associated taxa are clustered together is shown next to the branches of the phylogenetic tree (Figure 3). Using universal primers ITS1 and ITS4A, a total of 9 out of 18 mangrove fungal isolates were successfully amplified. BLAST searches revealed the identities of the mangrove fungal isolates as members of one genus, Aspergillus, and the phylum Ascomycota. Aspergillus flavus, Aspergillus niger, Aspergillus nomius, Aspergillus tubingensis, Aspergillus oryzae, Aspergillus awamori, Aspergillus aculeatus, Aspergillus brunneoviolaceus, and Aspergillus welwitchiae. The genus Aspergillus dominated in this study accounting for 50% of the isolates identified (Figure 3).
Table 1. Identification of mangrove fungal isolates from coastal Kenya using NCBI BLAST.
Preliminary screening
Preliminary screening of mangrove fungal extracts by paper disk diffusion showed that 4 out of 18 fungal endophytes inhibited the growth of test pathogens, gram-positive Staphylococcus aureus and gram-negative Escherichia coli. The zones of inhibition recorded at 50 mg/ml were between 5.67 and 17.1 mm. Methanolic extracts of isolates SaT2 and RmM4 inhibited both S. aureus and E. coli growth (Table 2). Methanolic extracts of isolates SaT1 and AmT2 only inhibited S. aureus (9.2 ± 0.75) and (6.16 ± 4.36), respectively. Figure 4 showed that ethyl acetate crude extracts of SaT1 exhibited the highest activity against both S. aureus (17.1 ± 2.45) and E. coli (15.9 ± 2.45). For all the extracts, the inhibition diameters of the fungal extracts were lower (5.67-17.1mm) than those of the positive control (24.77 ± 1.26). Results of 1-way ANOVA showed no significant difference in inhibition zone diameters between S. aureus and E. coli (df = 1; f = 2.182; p = 0.151)
The values of the means of inhibition diameters appended by different superscripts along a column were significantly different (p < 0.05). For instance, 9.2±0.75a significantly differed from 6.16±4.36b. The values of means whose inhibition diameters within respective columns are followed by a superscript of a similar lower-case letter such as a and ac or c and ac were not significantly different from each other (p > 0.05). Means with different superscripts along the same row are significantly different (p < 0.05) (Table 2).
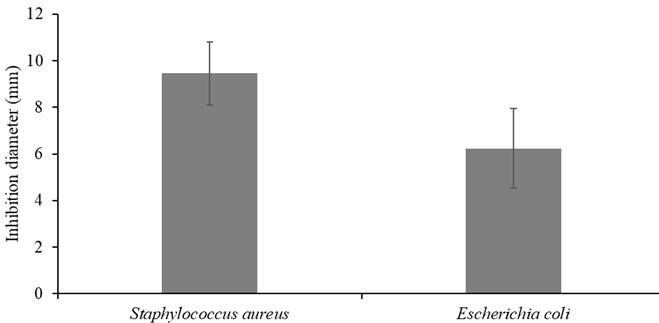
Table 2. In vitro antibacterial susceptibility tests of crude extracts of mangrove fungal endophytes (Mangrove fungal endophytes from coastal Kenya).
Determination of minimum inhibition concentration
The minimum inhibition concentration (MIC) value for E. coli was higher (0.91 ± 0.05 mg/ml) than that of S. aureus (0.82 ± 0.05 mg/ml) (Table 3). This implies that S. aureus was more susceptible to inhibition by active secondary metabolites of mangrove endophytic fungi than E. coli. The MIC values of the positive control for both test pathogens were higher than those of the test pathogens, ranging from 1.05 ± 0.10 to 1.08 ± 0.10 mg/ml for both Gram-positive S. aureus (ATCC 25922) and Gram-negative E. coli (ATCC 25922), respectively. The MIC value of E. coli (0.91 ± 0.05) was higher than that of S. aureus (0.82 ± 0.052).
Table 3. Minimum inhibition concentration of ethyl acetate crude extracts of Aspergillus flavus (Mangrove fungal endophytes from coastal Kenya).
DISCUSSION
A previous study isolated and identified the mangrove fungal endophytes from the twigs of mangrove species of Rhizophora mucronata, Avicenia marina, Ceriops tagal, and Sonneratia alba collected from coastal Kenya [14]. ITS sequences identified 9 isolates (50%) from the genus Aspergillus belonging to the phylum Ascomycota as observed in this present study. In a similar study [21], a total of 78 fungal isolates were identified from ITS sequences, 75 of which represented the phylum Ascomycota and only 3 of the phylum Basidiomycota. The phylum Ascomycota is said to be the most common representative of endophytic fungal communities isolated using standard procedures. It has also been reported that only a small portion of microorganisms in nature are culturable using standard microbiological techniques. This is because factors unfavorable for the growth of microorganisms exist in most laboratories. Some of the factors include limited knowledge of appropriate nutritive requirements and the challenging nature of microbes, especially when they are obtained from environmental samples [21]. These findings are in agreement with the findings of the present study, where 9 (50%) of the 18 fungal isolates were fully identified as belonging to the genus Aspergillus in the family Ascomycota. It is thought that laboratory conditions may have interfered with the growth of the fugal isolates, only favoring the growth of Aspergillus sp. [21]. Aspergillus flavus isolated from the mangrove plant Avicenia officinalis from Matang Mangrove Forest Reserve (MMFR), Perak, Malaysia was found to possess antioxidant properties [21]. All the fungal isolates identified except isolate CtT1 and AmM2 identified as Aspergillus aculeatus and Aspergillus brunneoviolaceus showed a similarity of 98.81% and 98.93%, respectively, above the percentage identity threshold set at about 97.5%.
Mangrove fungal endophytes have been a source of biologically active secondary metabolites with unique physiological and biochemical properties [9, 26]. In the current study, both ethyl acetate and methanolic crude extracts of fungal endophytes from stem twigs of selected mangrove species of coastal Kenya showed significant resistance against standard strains of S. aureus and E. coli. This suggests that crude extracts of mangrove fungal endophytes contain biologically active compounds inhibiting the growth of both gram-positive S. aureus (ATCC 25922) and gram-negative E. coli (ATCC 25922) [21] has reported the antibacterial activities exhibited by mangrove fungal extracts from the leaves of Rhizophora mucronata and Sonneratia caseolaris. In a similar study of antibacterial activities, ethyl acetate leaf extracts of different parts of Avicenia marina showed antibacterial activity against S. aureus and E. coli [1]. The activity of mangrove endophytic fungal extracts was attributed to the presence of secondary metabolites such as alkaloids, flavonoids, quinones, and terpenoids. These extracts showed significant inhibition against multidrug pathogenic bacteria, S. aureus [21]. In each case, inhibition diameters for both ethyl acetate and methanolic extracts were higher for S. aureus than for E. coli. This implies that the antibacterial activities of the fungal isolates were more common against the gram-positive S. aureus than the gram-negative E. coli and that S. aureus was more susceptible to the endophytic fungal extracts than E. coli. This observation was also reported [27]. These authors stated that the higher resistance of gram-negative bacteria E. coli was attributed to the presence of an outer double membrane surrounding the cells, which prevents certain drugs and bioactive/antimicrobial compounds from penetrating the cells, thereby making E. coli more resistant than S. aureus. Generally, ethyl acetate crude extracts exhibited higher inhibition diameters than methanolic extracts. For instance, ethyl acetate crude extract of isolate SaT1 identified as Aspergillus flavus showed the highest activity against S. aureus (ATCC 25922) (17.1 mm) and E. coli (ATCC 25922) (15.9 mm) as compared to methanolic extracts, recording a maximum inhibition of 10.0 mm against S. aureus and 8.96 mm against E. coli at 50 g/ml.
The zones of inhibition of ethyl acetate extracts of Avicenia marina leaves by an Aspergillus sp. ranged from 8.33 to 10.67 mm at concentrations ranging from 60 to 100 mg/ml, according to [27]. This is because leaf extracts are reported to contain more fungal metabolites than the stem twigs [28]. A study [10] observed that ethyl acetate is preferred in the extraction of fungal metabolites because it is able to extract both extracellular fungal metabolites (excreted by fungi into the medium) and intracellular bioactive compounds, an observation that is in agreement with the findings of the current study. According to a study [18], ethyl acetate is commonly used for the extraction of mangrove fungal metabolites since it is a medium-polar solvent with the ability to dissolve both polar and non-polar active compounds. This may imply that the most active secondary metabolites are medium polar and extractable by ethyl acetate. Methanolic crude extracts of all the fungal isolates showed activity against S. aureus. This implies that methanol was effective in extracting bioactive compounds from all the fungal endophytes that were active against S. aureus compared to ethyl acetate, an indicator that the activity of fungal extracts in this study was mainly as a result of polar active compounds extractable by methanol. According to a study [18], because methanol is polar, it can dissolve almost all organic compounds, including polar, semi-polar, and non-polar compounds. Aspergillus sp. has been shown to be effective in inhibiting microbial growth [21]. In the present study, the most active fungal endophyte was found to be SaT1 Aspergillus flavus isolated from A. marina of Mida Creek, north coast Kenya [14].
In conclusion, the present study has revealed that selected mangrove species of coastal Kenya are rich in endophytic fungi which produce antibacterial substances. A total of 9 isolates were identified as belonging to one phylum, Ascomycota, and represented only one genus, Aspergillus sp. Two fungal species, A. aculeatus (MZ314732) and A. brunneoviolaceus (MZ314735), were identified with up to 98.81% and 98.93%, respectively, an observation that could suggest a need for further investigation. The findings of this study also confirm that endophytic fungi from harsh environments such as mangrove ecosystems are an attractive source of biologically active compounds inhibiting the growth of both gram-positive S. aureus (ATCC 25922) and gram-negative E. coli (ATCC 25922). Thus, ethyl acetate crude extract of Aspergillus flavus showed greater activity against both test pathogens as compared to methanolic crude extracts.
Ethyl acetate and methanol were used as extracting solvents in this study. The study recommends the use of other extracting solvents and subsequent tests against S. aureus (ATCC 25922) and E. coli. (ATCC 25922). This study also recommends further investigation of the rest of the mangrove fungal extracts for activity against a wider range of pathogenic microbes, which are largely underexplored in this region. Two of the fungal endophytes, A. aculeatus (MZ314732) and A. brunneoviolaceus, (MZ314735) were identified up to 98.81 and 98.93 (%), respectively, were not fully identified indicative of the need for further research on these two isolates. The isolation of the specific chemical agents responsible for activity against bacteria is hereby recommended for drug development and manufacture.
ACKNOWLEDGEMENTS
The authors of this study appreciate the Technical University of Mombasa (TUM) and Kenya Marine and Fisheries Institute (KMFRI) for providing technical support and laboratory space over the study period. We also acknowledge Alexander Fulanda and Mackmillan for support in the isolation, purification, microscopy, antimicrobial assays, and morphological and molecular characterization of mangrove fungal endophytes at KMFRI laboratories. This study was partly funded by a marine research grant from the Western Indian Ocean Marine Science Association (WIOMSA) and the National Research Fund (NRF). It was also approved by the National Commission for Science, Technology, and Innovation (NACOSTI), (approval number: TUM ERC PHD/002/2020). This work forms part of the requirement for the Doctor of Philosophy Degree at TUM by the first author.
AUTHORS CONTRIBUTION
HMK: Funding acquisition, Conceptualization, methodology, fieldwork, laboratory work, analysis and writing of original draft. CMK: Methodology, supervision, data analysis and editing; CNM: Funding acquisition, supervision, data analysis and editing; JOO: Fund acquisition, conceptualization, methodology, supervision and editing; PMG: Conceptualization, methodology, supervision and editing, and SMM: laboratory work and data analysis. All authors approved the final manuscript for publication.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Okla MK, Alatar AA, Al-Amri SS, Soufan WH, Ahmad A, Abdel-Maksoud MA. Antibacterial and Antifungal Activity of the Extracts of Different Parts of Avicennia marina (Forssk.) Vierh. Plants (Basel). 2021; 10(2).
- [2]Organization WH. Antimicrobial resistance. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance, 2021.
- [3]aranraj PSD, Mangrove Medicinal Plants: A Review. American-Eurasian Journal of Toxicological Sciences, 2015; 7(3): 146-156.
- [4]Dadar M, Tiwari R, Karthik K, Chakraborty S, Shahali Y, Dhama K. Candida albicans – Biology, molecular characterization, pathogenicity, and advances in diagnosis and control – An update. Microbial pathogenesis, 2018; 117: 128-138.
- [5]eoh WP, Chen X, Laczkovich I, Alonzo F. Staphylococcus aureus adapts to the host nutritional landscape to overcome tissue-specific branched-chain fatty acid requirement. Proceedings of the National Academy of Sciences of the United States of America, 2021; 118(13).
- [6]Chigodi MO. Isolation and screening of Antipneumonic compounds from Acacia Stuhlmannii Taub. (Master of Science in Chemistry pdf), Pwani University, 2017.
- [7]Huttanus HM, Sheng J, Feng X. Metabolic Engineering for Production of Small Molecule Drugs: Challenges and Solutions. Fermentation, 2016; 2(1): 4.
- [8]Bijaya KN. Studies on endophytic fungal diversity from different leaf samples of Pongamia pinnata. International Journal of MediPharm Research, 2015; 01(02): 134-138.
- [9]Cadamuro RD, da Silveira Bastos IMA, Silva IT, da Cruz ACC, Robl D, Sandjo LP, Alves SJr, Lorenzo J M, Rodriguez-Lazaro D, Treichel H, Steindel M, Fongaro G. Bioactive Compounds from Mangrove Endophytic Fungus and Their Uses for Microorganism Control. Journal of Fungi (Basel), 2021; 7(6).
- [10]Ling MO, Teen PL, Mujahidin A, Proksch P, Muller M. Initial Screening of Mangrove Endophytic Fungi for Antimicrobial Compounds and Heavy Metal Biosorption Potential. Sains Malaysiana, 2016; 45(7): 1063–1071.
- [11]Ling M, Onn L, Teen P, Aazani M, Peter P, Moritz, M. Initial screening of mangrove endophytic fungi for antimicrobial compounds and heavy metal biosorption potential. University of Malaya, 2013.
- [12]Programme, U. N. E. (2014). The importance of mangroves to people: a call to action. https://wedocs.unep.org/20.500.11822/9300.
- [13]Dahdouh-Guebas F, Mathenge C, Kairo JG, Koedam N. Utilization of mangrove wood products around Mida creek (Kenya) amongst subsistence and commercial users. Economic Botany, 2000; 54(4): 513-527.
- [14]Kiti HM, Munga CM, Odalo JO, Guyo PM, Kibiti CM. Diversity of mangrove fungal endophytes from selected mangrove species of coastal Kenya. Western Indian Ocean Journal of Marine Science, 2021; 20(1): 125-136.
- [15]Kairo JG, Dahdouh-Guebas F, Bosire J, Koedam N. Restoration and management of mangrove systems — a lesson for and from the East African region. South African Journal of Botany, 2001; 67(3): 383-389.
- [16]Jenoh EM, de Villiers EP, de Villiers SM, Okoth S, Jefwa J, Kioko E, Kaimenyi D, Hendrickx M, Dahdouh-Guebas F, Koedam N. Infestation mechanisms of two woodborer species in the mangrove Sonneratia alba J. Smith in Kenya and co-occurring endophytic fungi. PLOS ONE, 2019; 14(10): e0221285.
- [17]Bosire JO, Kaino JJ, Olagoke AO, Mwihaki LM, Ogendi GM, Kairo JG, Berger U, Macharia D. Mangroves in peril: unprecedented degradation rates of peri-urban mangroves in Kenya. Biogeosciences, 2014; 11(10): 2623-2634.
- [18]Nurunnabi T, Al-Majmaie S, Nakouti I, Nahar L, Rahman S, Sohrab M, Billah M, Ismail F, Sharples G, Sarker S. Antimicrobial activity of kojic acid from endophytic fungus Colletotrichum gloeosporioides isolated from Sonneratia apetala, a mangrove plant of the Sundarbans. Asian Pacific Journal of Tropical Medicine, 2018; 11(5): 350-354.
- [19]Kibiti CM, Afolayan AJ. Preliminary Phytochemical Screening and Biological Activities of Bulbine abyssinica Used in the Folk Medicine in the Eastern Cape Province, South Africa. Evidence-based complementary and alternative medicine: eCAM, 2015; 617607-617607.
- [20]Moron LS, Lim YW, Cruz TE. Antimicrobial activities of crude culture extracts from mangrove fungal endophytes collected in Luzon Island, Philippines, 2018.
- [21]Hamzah TNT, Lee SY, Hidayat A, Terhem R, Faridah-Hanum I, Mohamed R. Diversity and Characterization of Endophytic Fungi Isolated from the Tropical Mangrove Species, Rhizophora mucronata, and Identification of Potential Antagonists Against the Soil-Borne Fungus, Fusarium solani. Front Microbial, 2018; 9: 1707.
- [22]Thorati MMJK, Kumar S. Isolation, Identification of Endophytic Fungi from Mangrove Roots along the Coast of South Andaman Sea, Andaman and Nicobar Islands, India. Journal of Marine Biology & Oceanography, 2016; 5(2): 1-5.
- [23]Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 1980; 16(2): 111-120.
- [24]Kumar S, Kaushik N, Proksch P. Identification of antifungal principle in the solvent extract of an endophytic fungus Chaetomium globosum from Withania somnifera. SpringerPlus, 2013; 2(1): 37.
- [25]Mao Z, Zhang W, Wu C, Feng H, Peng Y, Shahid H, Cui Z, Ding P, Shan T. Diversity and antibacterial activity of fungal endophytes from Eucalyptus exserta. BMC Microbial, 2021; 21(1): 155.
- [26]Ababutain IM, Aldosary SK, Aljuraifani AA, Alghamdi AI, Alabdalall AH, Al-Khaldi EM, Aldakeel SA, Almandil NB, AbdulAzeez S, Borgio JF. Identification and Antibacterial Characterization of Endophytic Fungi from Artemisia sieberi. International journal of microbiology, 2021; 6651020-6651020.
- [27]Ariole CN, Akinduyite AE. Antibacterial potential of indigenous red mangrove (Rhizophora racemosa) fungal endophytes and bioactive compounds identification. International Network for Natural Sciences, 2016; 4: 14-24.
- [28]Ariole CN, Akinduyite, AE. Bioactive compounds and antimicrobial activity of endophytic fungi isolated from Black Mangrove (Avicenia Africana) leaves. Nigerian. Journal of Biotechnology, 2018; 35(2): 35 – 42.