Isolation, documentation, and biochemical characterization of cellulolytic bacteria from rumen fluid of cattle
Abstract
The microbiological use of cellulose is a significant aspect in achieving the highest possible material flow in the environment. This study aimed at isolation, identification, and biochemical characterization of bacteria with cellulase activity from cellulose samples. Cellulase enzyme has wide applications in various industries including food industry, agriculture, textile, detergent, pulp and paper, biofuel production, brewing and biorefinery. In the present study, cellulolytic bacterium Pseudomonas sp. was isolated from rumen fluid of cattle was collected from slaughter house. This bacterial isolate was identified by morphological, biochemical, and physiochemical characteristics. Cellulase production by the bacteria was optimized. Optimum cellulase was produced by Pseudomonas sp. which was observed under different pH, temperature, and incubation period. The highest production of cellulase enzyme by this bacterium was monitored at pH 7.0 for 48 hours under 3 days of cultivation at 40°C. The enzyme activity was observed at pH 7.0 with cellulosic filter paper for 5 days incubation and its maximum activity was noted at 48 hours. The result of study shows that the Pseudomonas sp. is a good producer of extra cellular cellulase enzyme which can be also beneficial for the degradation of cellulosic pollutants of environment.
INTRODUCTION
Cellulose is most abundant element of plant biomass and agricultural waste is considered as low-cost biopolymer and renewable energy source [1]. Plant polysaccharide cellulose is primarily fibrous crystalline in nature and consists of the repeating units of β-D-glucose monomers, linked by β-1, 4- glycosidic bond having high molecular weight and soluble in water [2, 3]. This can be broken down into glucose monomers and into other soluble sugars by cellulolysis process. Cellulolysis process is triggered by a set of enzymes called cellulase, which comprises of endoglucanase (endo-1, 4-β-D-glucanase), exoglucanase (exo-1, 4-β-Dglucanase) and β-glucosidase (1, 4-β-D-glucosidase) [4]. According to Cheng et al., every year more than hundreds (100) billion metric tons of cellulose is found naturally, whereas the total biomass is about two hundred eighty (280) billion metric tons [5].
Cellulase is well suited for a variety of agro-industrial processes that use biofuels such as bioethanol [6], management of plant and agricultural waste, binding of ligand and separation of chiral studies, thereby resulting in upgrading of the waste or the synthesis of valuable by-products [7]. But there is a shortage of microorganisms those are capable of producing large amounts of cellulase enzymes toward proficiently convert cellulose into fermentable products [8]. Numerous microorganisms synthesize cellulose, a bioactive compound for the period of their life span on cellulosic materials [9]. In soils, cellulosic compounds are degraded mostly by cellulolytic microorganisms such as fungi and bacteria [10]. Bacteria are currently extensively studied for cellulose production due to their enormously high natural diversity and ability to produce stable enzymes that can be used in industry [11, 12]. The prospective cellulose synthesizing bacteria are Cellulomonas, Pseudomonas, Thermoactinomycetes, Bacillus sp. Clostridium, Cellulomonas, Cellulosimicrobium, Thermomonospora [13]. Bacterial cellulase generally act as effective and potent catalyst [14] which is extensively used for their rapid growth, multi-enzyme complexes expression. Moreover, their stability at high temperature and pH helps to reduce activity of inhibition and ability to colonize a comprehensive range of environmental niches [15, 8]. Additionally, nature and quality of cellulose, temperature, pH of the medium, incubation period, carbon sources, medium additives, and existence of diverse inducers are also important parameters for the maximization of different cellulase enzymes production [16].
Research on the isolation and characterization of vigorous cellulase producing microorganisms from different sources have been sustained for numerous years [17]. Most of the previous studies emphasized more on fungi than on bacteria as a potential source for the production of cellulase. Therefore, the primary objective of this research was the isolation and identification of cellulytic bacteria from rumen fluid, their ability to produce cellulase enzyme and the effect of pH, temperature, and incubation period in the production of cellulase enzyme.
MATERIALS AND METHODS
Sample collection
Rumen fluid samples were collected from slaughtering cattle at Kajitula Slaughtering house, Sylhet City Corporation, Bangladesh. Flasks containing warm water were used to carry out rumen fluid following Khan and Chaudhry, [18] protocol in order to maintain survival temperature for microbes. The collected liquid was squeezed out, filtered through a 4-layer cheesecloth, sealed, and stored at 39 ° C.
Reagents and culture media preparation
Analytically graded inorganic reagents KH2PO4 1.0 g, K2HPO4 1.145 g, MgSO4 0.4 g, NH4SO4 5.0 g, CaCl2 0.05 g, FeSO4 0.00125 g, carboxy methyl cellulose (CMC) 10.0 g and agar 18g were dissolved in 1000 ml double distilled water (ddH2O) to prepare culture media. The prepared media was then sterilized by autoclaving at 121°C, 15 psi for around 30 min and subsequently introduced into sterilized petri dishes that were oven dried at 180 °C for 1 hour.
Isolation of cellulolytic bacteria
At first, rumen fluid (1 ml) was diluted with 9 ml sterile double distilled water (ddH2O) (v/v) where 10−1 to 10−5 serial dilutions were prepared. Then 0.3 ml of samples was taken from each dilution and streaked into petri dishes containing CMC medium and incubated at 37°C for 24 hours [31]. After 24 h of incubation, no clear zone was formed around the bacterial culture that’s why it was applied iodine to visualize the clear zones produced by cellulolytic bacteria. Subsequently, well grown colonies were chosen to obtain a pure culture of bacteria using two loops of an inoculating needle to transfer and inoculate onto a petri dish containing CMC medium [19]. The selection of cellulolytic species was conducted based on the ratio of clear zone to colony diameter after 48 hours on carboxy methyl cellulose (CMC) media. Later, subcultures were made to produce new culture with a lower density of cells.
No ethical approval is needed here as rumen fluid was collected from rumen after normal slaughtering of cattle.
Characterization of cellulose-producing bacteria microscopically with gram staining
Gram stain was an empirical method of distinguishing bacterial species into two large groups (Gram-positive and Gram-negative) based on the presence of chemicals, primarily the presence of high levels of peptidoglycan and physical properties of their cell walls. A small colony was taken using a loop needle, then smeared on a glass slide and fixed by gentle heating over Bunsen. Gram A (Crystal violet- primary stain) solution was then applied on the smear to stain and left for 2 minutes then rinsed with running water. Next, dropped with Gram B (Lugol’s iodine) solution as mordant for 1 minute and then washed with running water. Then 95 % ethanol was added until the remaining dye disappears for 5 seconds and rinsed again with running water. In the final stage, the preparation was dropped with gram C (safranin) solution as counter stain and allowed to stain and dry for 45 seconds. Then it was drizzled with immersion oil and examined under a microscope at a magnification of 100X. After that, the slide was washed, blotted, and dried in the air, and the observations were performed by looking at cell morphology and color. Gram-positive bacteria change color to purple or blue, while Gram-negative bacteria change color to red [20].
Biochemical test
Catalase test
The catalase test was performed according to Cappucino and Sherman’s method [19]. Colonies that had been cultured for 18-24 hours were transferred to a glass slide with a loop, and 1-2 drops of 3 percent hydrogen peroxide were applied to the colonies. The presence of gas bubbles in the glass slide indicated positive results, whereas the absence of gas bubbles indicated catalase negative results [21].
Oxidase test
Oxidase test was carried out by smearing isolates in filter paper that was soaked with tetramethyl-p-phenylenediaminedihydrochloride. Inoculated were observed on paper for a color change to deep blue or purple within 10-30 seconds. Violet color indicates oxidase positive and white color indicates oxidase negative [21].
Citrate test
Inoculate Simmons citrate agar on the slant by touching a colony that was 18-24 h old with a straight wire. Stabbing the butt and broth culture of the medium was strictly prohibited. After 7 days incubation at 35-37°C, bacterial growth with color changes from green to intense blue along with the slant indicates citrate positive and green slant with no color change indicates citrate negative [21].
Indole test
Indole test was a commonly used biochemical test to differentiate Enterobacteriaceae and other genera. Indole test was used to determine the ability of an organism to split amino acid tryptophan to form the compound indole. Indole production was detected by Kovac’s or Ehrlich’s reagent which contains 4(p)-dimethylamino benzaldehyde, this reacts with indole to produce a pink or red colored compound indicate indole positive and no color change indicate indole negative [21].
Methyl red test
Unknown microorganism was inoculated by using sterile inoculation loop into sterile medium, which was prepared from mixing of 7g buffered peptone, 5g glucose and 5g Di-potasium phosphate in per liter of deionized water. The inoculated tube was then subjected to incubate at 35-37°C for two to five days. After incubation, the broths were obtained from the incubator and 5 drops of Methyl Red reagent was added to the broth. Color changes to red indicate Methyl red positive and no color changes indicate Methyl red negative [21].
Voges-Proskauer test
A tube of MR/VP broth (reagents of alpha-napthol (5%) and absolute ethanol) was taken and a pure culture of the test organism was inoculated into it. Then it was incubated for 24 to 48 hours at 35°C. At the end of this time, aliquot of 1 ml brought to clean test tube. A 0.6 ml of 5% alpha naphthol was added followed by 0.2 ml of 40 % KOH. To expose the medium to atmospheric oxygen, gentle shaking of the tube gently was practiced and then allowed to remain undisturbed for 10 to 15 min. A pink-red color at the surface indicated positive result for this test [21].
Glucose fermentation test
At first, trypticase 3g, sodium chloride 5g and phenol red 0.018g were weighed and dissolved in 100 ml distilled water and transferred into conical flasks. Then, 0.5% of dextrose was added into the flasks and autoclaved at 115oC for 15 minutes. The mixture was then transferred into fermentation tubes and labeled properly. Carbohydrate broths were inoculated with aseptically labeled bacterial culture and incubated the tubes for 18-24 hours at 37oC. Color changes to yellow indicated positive and no color changes indicated negative [21].
Cellulolytic enzyme production and assay
For cellulolytic enzyme production, at first seed production in CMC media (40 ml) was prepared without agar in a 100 ml conical flask. Later, 10 ml of CMC solution was poured into three conical flasks and maintained different pH (6.0, 6.5 and 7.0) which was measured and set accordingly. Then it was autoclaved and inoculated with 1% inoculum (v/v) using sterilized inoculating loop. The preparation was then subjected to incubate in a shaking incubator at 37°C at 120 rpm for 24 hours. The media without the culture was considered as control. After 18 hours enzyme was collected and evaluated it.
Screening for enzymes production
At the end of the incubation, the agar medium along with bacterial colonies was flooded with an aqueous solution of Congo red (1% w/v) for 15 minutes. The congo red solution was then poured off and the plates were further treated by flooding with 1M NaCl for 15 minutes. The formation of a clear zone due to hydrolysis, indicated cellulose degradation, but the results were so poor that Gram’s iodine (KI+I2) was used [22]. The ratio of the highest cellulose activity producer was assumed to contain the highest activity [23, 24].
Cellulolytic enzyme activity assay
Filter paper (FPase) was used to measure the activity of cellulolytic enzyme, as described by Mandels and Weber [25]. 1.8 ml 50 micromole sodium citrate buffer was absorbed with double ring 102 filter paper (pH 4.8). Then 0.2 ml of culture was added and incubated for 60 minutes at 40 0C. The reaction was stopped by adding 3.0 ml of dinitro-salicylic acid (DNS) reagent to the reagent tubes and placing them in a water bath at 100 °C for 15 minutes. To stabilize the color, 1 ml Rochelle salt solution (40 g Rochelle salt in 100 ml distilled water) was added. In a spectrophotometer, the absorbance/optical density (OD) was measured at 575 nm against a blank of 50 µM citrate buffer. One micromole of glucose liberated per ml enzyme per minute equaled one unit of CMCase activity.
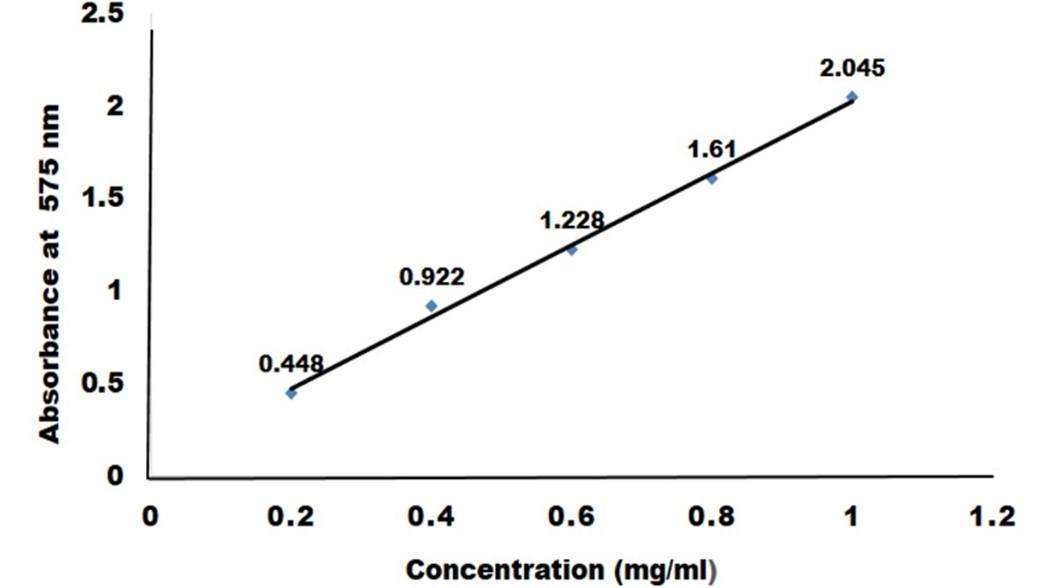
Measurement of glucose liberated by crude cellulase
To create a standard curve, the absorbance of a known glucose concentration solution (0.2 to 1 mg/ml) was measured in a spectrophotometer and compared to distilled water (control) as mentioned above. 1 µM of glucose liberated per ml enzyme per minute was defined as one unit of enzyme.
Effect of pH on the cellulase production
The influence of optimum pH for cellulase production by the experimental microorganism was determined by culturing the bacteria and measuring the enzyme activity at different pH value ranging from 6, 6.5 and 7 at optimum condition. Different suitable buffer, 50mM sodium citrate (pH 4.0), 50 mM sodium acetate (pH 5.0 and 6.0), 50 mM sodium phosphate (pH 7.0 and 8.0) were used.
Effect of temperature on the cellulase production
Temperature plays a significant role for the production of cellulase by the test organism. To observe the effect of temperature on the production of cellulase, the fermentation was carried out at different temperatures ranging from 25-45°C.
Effect of incubation period on cellulase activity
The experiment was carried out to find out the effect of incubation time on the production of cellulose. The flasks containing 50 ml sterile production medium were inoculated with 10 ml seed culture and incubated for 3 days at optimum temperature.
RESULTS
Isolation and screening of cellulolytic bacteria from rumen fluid sample
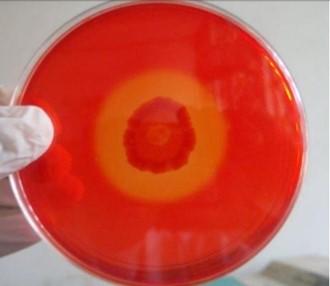
A number of cellulolytic bacterial colonies were isolated from the rumen fluid of cattle on CMC medium and screening of cellulolytic activity were visualized using Congo red staining 0.1%. Degradation of cellulose was indicated by the presence of a clear zone around the bacterial colonies as shown in Figure 2. Strain was found to have cellulolytic activity and based on the cellulolytic index calculated by the diameter of the clear zone; the isolate had cellulolytic activity. +2.5 cm (Figure 2). Clear zone producing bacterial isolates were then subjected to various biochemical tests [26] .
Biochemical test for genus identification
Results from the biochemical tests, it was observed that the bacterium was a gram-negative and rod shaped. Bubbles of oxygen gas were generated on the slide when the amylolytic bacterial colonies were brought for catalase test that indicates the isolates have the ability to catalyze H2O2. Enzyme Catalase catalyzes the breakdown of H2O2 and O2. Hydrogen peroxide (H2O2) is toxic to cells and so it stimulates enzymes in cells. In case of oxidase test, the colonies showed positive results by producing purple color on moist filter paper that is able to produce cytochrome C oxidase (Table 1). The other genus confirmatory identification test was based on methyl red, voges-proskuer, glucose fermentation, citrate test and indole test. Among these, the bacterial colonies gave positive result for methyl red, voges-proskuer, glucose fermentation, citrate test except for indole test (Table 1). Based on biochemical and morphological test results, the isolates were identified to belong to the genus of Pseudomonas sp. This bacterial genus is gram negative; rod shaped and have cellulolytic activities.
Table 1. Biochemical test for identification of Genus.
Effect of pH on cellulase production
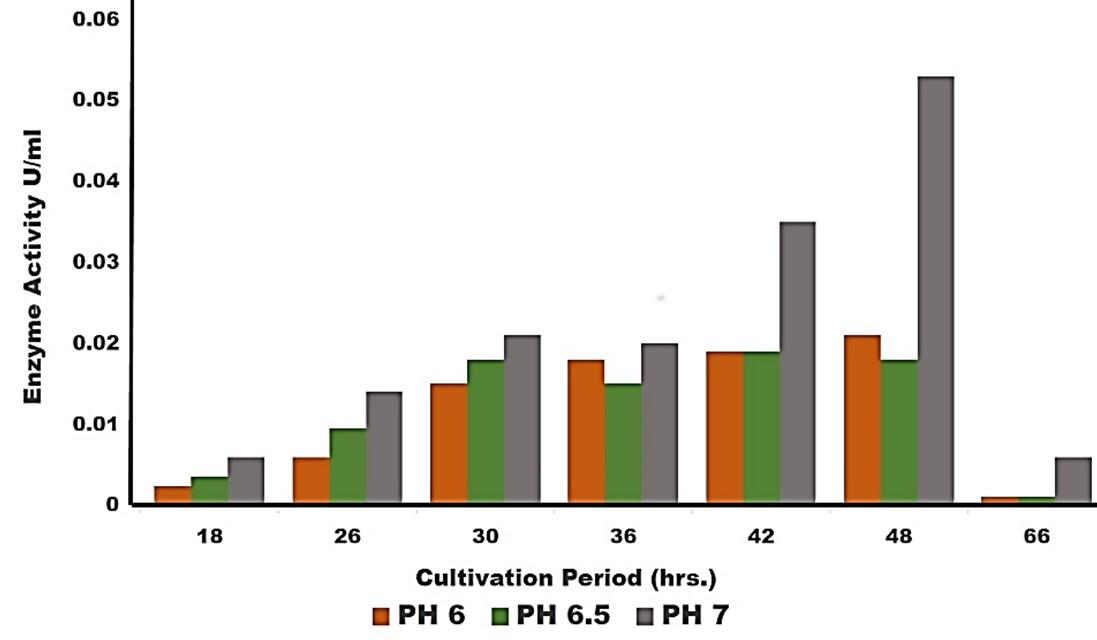
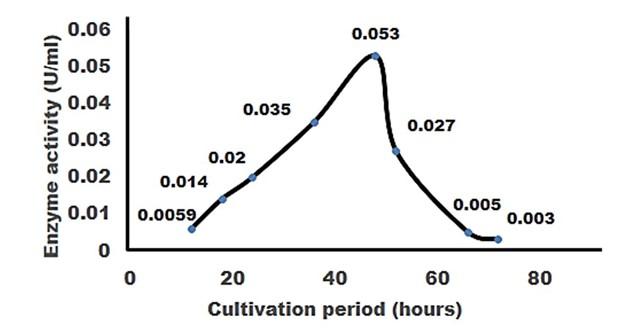
Cellulase production by the Pseudomonas sp. isolated from rumen fluid of cattle was adjusted under varying cultural conditions and observed at different pH i.e. 6, 6.5, 7 after 72 hours of incubation at 37°C (Figure 3). Maximum cellulase production was recorded at pH 7.0 (0.053 U/ml) and the minimum cellulase production was recorded at pH 6 and 6.5 (0.0011 U/ml) after 66 hours of cultivation period.
Effect of temperature on cellulase production
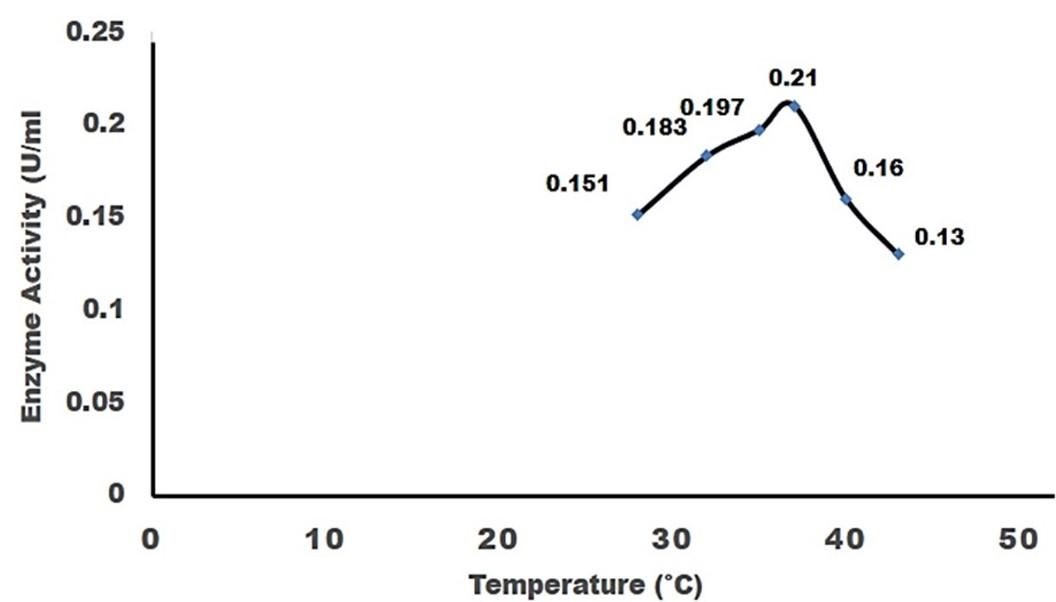
For the production of cellulase enzyme, the identified bacteria (Pseudomonas sp.) was taken under different temperature 28°C, 32°C, 35° C, 37° C, 40° C and 43° C. Among them, the bacteria showed the highest level of cellulase production at 370 C (0.21 U/ml) (Figure 4).
Effect of incubation period on cellulose production
For producing cellulase enzyme, the identified bacteria (Pseudomonas sp.) was taken under different incubation period 12,18, 24, 36, 48, 52, 66 and 72 hours. Among them, the bacteria showed the maximum production (0.053 U/ml) at 48 hours of incubation period (Figure 5).
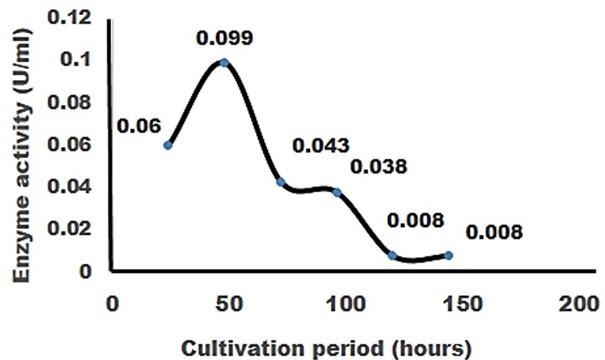
Effect of cellulase enzyme on filter paper
The result of the test was strong evident that cellulase was produced in order to degrade cellulose. CMC was most effective as a carbon source for cellulose enzyme production by Pseudomonus sp. result in increase in enzyme activity [27]. The application of cellulase enzyme was carried by using double ring 102 filter paper. It was observed that the cellulase enzyme showed maximum degradation in 2nd day under 5 days of incubation in shaking incubator (Figure 6).
In short, it was found that isolated bacterium was Pseudomonus sp. Where CMC used as the production media. This bacterium produced optimum level of cellulase at 37°C, maximum production occurred at 48 hours of incubation period at pH 7 and filter paper degradation was highest at 2nd day of incubation.
DISCUSSION
As cellulose is abundant in nature and awaited to be converted into more valuable products used for mankind. Numerous microorganisms capable of converting polysaccharide cellulose into simple carbohydrates had been revealed for decades. Cellulase enzyme that is commonly used in several industries, the present study was dealt with isolation, identification and biochemical characterization of cellulolytic bacteria and assessed their potentiality. The cellulose-containing rumen fluid of cattle samples was used for this study, where only one bacterial isolates were identified with higher enzymatic activities, cellulose degraders and considered to use as cellulose degradation to decompose plants more effectively [28].
CMC media is utilized for cellulose production as well as the identification and screening of cellulolytic bacteria by the formation of a clear hydrolytic zone [29, 30]. When CMC is used as a substrate, more cellulose is produced, which could be due to the enzyme being introduced, as cellulose is known to be a universal inducer of cellulase synthesis.
Pseudomonas sp., a cellulolytic bacterium, was isolated from cattle rumen fluid. This bacterial isolate was identified by morphological, biochemical, and physiochemical characteristics. Biochemical tests have been carried for the identification of unknown genus as described by Cowan and Steel [31]. In the present study cellulase production by the bacteria was observed. The major concern of present study is to keep an eye on the cellulase manufacturing circumstances. The most prominent features for optimizing enzyme production, according to Pelczar et al. [32], are pH and temperature. In the manufacturing of enzymes, the incubation duration is equally important. Pseudomonas sp. produced optimal cellulase at various pH, temperatures, and incubation times. Optimum temperature for this study was 40°C but a considerable amount of cellulase is produced from 35°C. Sohag et al. [33] and Khatiwada et al. [34] also found that optimum temperature was 40°C for Pseudomonas sp. It was previously reported that cellulolytic bacteria produced highest level of cellulase at a pH range of 6.0-7.5 [35, 36] and enzyme production through Pseudomonas sp. is required similar criteria and result found same as the previous study conducted by other researchers. The highest production of cellulase enzyme by Pseudomonas sp. was observed at pH 7.0 under 3 days of cultivation which is similar as the results of the experiment conducted by Sohag et al. [33] and Khatiwada et al. [34]. The highest enzyme activity was observed at pH 7.0 with cellulosic filter paper for 5 days of incubation and its maximum activity was noted at 48 hours. The result of study shows that the Pseudomonas sp. is a good producer of extra cellular enzyme cellulase which can be also beneficial for the degradation of cellulosic pollutants of environment.
The collection of more bacterial isolates from rumen fluid sample and genetic engineering approach would provide more pace to degrade the organic wastes that is now a concern for the development of ecologically sound and health promoting ways for the management of the environment.
CONCLUSIONS
Cellulase offers an important opportunity to bring great benefits to the use of biomass [37]. Cellulose-degrading microorganisms can alter cellulose into diverse soluble sugars by either enzymatic hydrolysis or acidic reaction. A significant amount of cellulase was produced by Pseudomonas sp. using cellulosic substrates in shake flask with optimized parameters. This bacterium is an important species and showed better cellulolytic activity. Besides, the crude enzyme isolated from Pseudomonas sp. acted efficiently for degradation of filter paper. So, it is expected that this bacterial species can be a good source to remove cellulosic pollutants from the environment. Using various techniques, in the case of cellulase, it can convey performance industrial application. In future, study of fungus can be isolated from rumen fluid and cellulolytic activity of that fungus could be monitored.
ACKNOWLEDGEMENT
This work is funded by Ministry of Science and Technology (MoST), Government of the People’s Republic of Bangladesh.
AUTHOR CONTRIBUTIONS
MMHK designed and supervised the overall research work; NYP and SM performed the research work; MNH wrote the manuscript and analyzed the data; AKA revised the manuscript and some of the lab works has conducted in his laboratory; MH critically overviewed the manuscript. All authors revised and approved the final version of the manuscript.
CONFLICTS OF INTEREST
There is no conflict of interest among the authors.
References
- [1]Khaleel HL, Abd AN, Ali KM. Preparation of Nano-Cellulose from Industrial Waste by Ultrasonic Device. Journal of Biochemical Technology 2018; 9: 35.
- [2]Zaghoud L, Gouamid M, Benmenine A, Khanblouche A. Kinetic and Thermodynamic of Gentian Violet Removal by 2, 3-Dialdehyde Nanocellulose. Journal of Biochemical Technology 2019; 10: 38-42.
- [3]Malherbe S, Cloete TE. Lignocellulose biodegradation: fundamentals and applications. Reviews in Environmental Sciences and Biotechnology 2003; 1: 105-114.
- [4]Li X, Gao P. Isolation and partial properties of cellulose-decomposing strain of Cytophaga sp. LX-7 from the soil. J. Appl. Microbiol. 2008; 82: 73-80.
- [5]Cheng Q, Wang J, McNeel JF, Jacobson PM. Water Retention Value Measurements of Cellulosic Materials Using a Centrifuge Technique. BioResources 2010; 5: 1945-1954.
- [6]Acharya A, Joshi DR, Shrestha K, Bhatta DR. Isolation and screening of thermophilic cellulolytic bacteria from compost piles. Scientific world. 2012; 10: 43-6.
- [7]Lu WJ, Wang HT, Nie YF. Effect of inoculating flower stalks and vegetable waste with lignocellulolytic microorganisms on the composting process. Journal of Environmental Science and Health 2004; 39: 871–887.
- [8]Maki M, Leung KT, Qin W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 2009; 5: 500-516.
- [9]Ojumu T, Solomon V, Bamidele O, Betiku E, Layokun SK. Cellulase Production by Aspergillus flavus Linn Isolate NSPR 101 fermented in sawdust, bagasse and corncob. African J. Biotechnol. 2003; 2: 150–152.
- [10]Gomashe AV, Gulhane PA, Bezalwar PM. Isolation and screening of cellulose degrading microbes from Nagpur region soil. International Journal of Life Science 2013; 1: 291-293.
- [11]Haakana H, Mittinen-Oinonen A, Joutsjoki V, Mantyla A, Souminen P, Vahmaanpera J. Cloning of cellulase from Melanocarpus albomyces and their efficient expression in Trichoderma reesei. Enzyme Microbial Technology 2004; 34: 159-167.
- [12]Ashjaran A, Sheybani S. Drug Release of Bacterial Cellulose as Antibacterial Nano Wound Dressing. International Journal of Pharmaceutical Research & Allied Sciences 2019; 8 :137-143.
- [13]Godana B, Mitra R, Singh S. Production of Enzymes for Application on Animal Feeds. Submitted in partial fulfillment of the requirements for the degree of Master of Technology (Biotechnology), Department of Biotechnology, Faculty of Science, Engineering and the Built Environment, Durban University of Technology, Durban, South Africa 2007.
- [14]Gautam SP, Bundela PS, Pandey AK, Awasthi MK, Sarsaiya S. Composting of municipal solid waste of Jabalpur City. Global Journal of Environmental Research 2010; 24: 43–46.
- [15]Sreeja SJ, Malar PWJ, Joseph FRS, Tiburcius S, Immanuel G, Palavesam A. Optimization of cellulase production by Bacillus altitudinis APS MSU and Bacillus licheniformis APS2 MSU, gut isolates of fish Etroplussuratensis. Int. J. Adv. Res. & Tech. 2013; 2: 401-406.
- [16]Angsana R, Warinthorn S, Annoop N, Pawinee C. Combination effect of pH and acetate on enzymatic cellulose hydrolysis. Journal of Environmental Science 2009; 21: 965- 970.
- [17]Doi RH. Cellulase of mesophilic microbes: cellulosome and non–cellulosome producers. Ann. N.Y. Acad. Sci. 2008; 1125: 267–279.
- [18]Khan MMH, Chaudhry AS. Comparing Rumen Fluid to Buffer Ratios to Estimate in vitro Degradability, Fermentation, and Methane Profiles of Seven Forages at Two Incubation Times. Iranian Journal of Applied Animal Science 2021; 11: 431-441.
- [19]Cappucino JG, Sherman N. Microbiology a Laboratory Mannual. 2th Ed. California: The Benjamins Columningn Publishers Company. 1987.
- [20]Faridha BI, Meignanalaksmi S, Pandima DM. Isolation and Characterization of Cellulase Producing Paracoccus Pantotrophus fmr19 (jx012237) from Goat Rumen Fluid and Its Effects on pH, Temperature and Carbon Sources. International Journal of Advanced Biotechnology and Research 2013; 4: 384-390.
- [21]Sharma N, Baliarsingh S, Kaushik GG. Serum Electrolytes Changes with Atherogenic Index of Plasma in Hypothyroid Hiabetic (type-2) Young Males. International Journal of Pharmaceutical Science and Research 2013; 4: 3046-3050.
- [22]Kasana R, et al. A rapid and easy method for the detection of microbial cellulases on agar plates using gram s iodine. Current Microbiology 2008; 57: 503-507.
- [23]Howard RL, Abotsi E, Jansen V, Rensburg EL, Howard S. Lignocellulose biotechnology: issues of bioconversion and enzyme production. Review: African Journal of Biotechnology 2003; 12: 602-619.
- [24]Ariffin H, Abdullah N, Hasan MA. Production and Characterization of Cellulase by Bacillus pumilus EB3, International Journal of Environmental Technology 2006; 3: 44-53.
- [25]Mandels M, Weber J. The production of celluases and their applications. Hajny, G.J. and E.T. Resse (ed.), Amer. Chem. Soc. Adv. Ser. 1969; 95: 391 -414.
- [26]Khatiwada P, Ahmed J, Sohag MH, Islam K, Azad AK. Isolation, screening and characterization of cellulase producing bacterial isolates from municipal solid wastes and rice straw wastes. J Bioprocess Biotech. 2016; 6: 2.
- [27]Gautam SP, Bundela PS, Pandey AK, Awasthi MK, Sarsaiya S. Composting of municipal solid waste of Jabalpur City. Global Journal of Environmental Research 2010; 24: 43–46.
- [28]Mahmood R, Afrin N, Jolly SN, Shilpi RY. Isolation and identification of cellulose-degrading bacteria from different types of samples. World 2020; 9: 8-13.
- [29]Das P, Solanki R, Khanna M. Isolation and screening of cellulolytic actinomycetes from diverse habitats. International Journal of Advanced Biotechnology and Research 2014; 5:4 38-451.
- [30]Gomashe AV, Gulhane PA, Bezalwar PM. Isolation and screening of cellulose degrading microbes from nagpur region soil. Int. J. of Life Sciences 2013; 1: 291-293.
- [31]Cowan I, Steel ST. Cowan and Steel’s Manual for the identification of medical bacteria.3rd edn. Cambridge University Press, London. 1974.
- [32]Pelczar JM, Chan ECS, Krieg RN. Microbiology. 5th edition 2004. Tata McGraw Hill, New Delhi, India.
- [33]Sohag MMH, Hasan MM, Ahmed J, Daud SNA, Alam MK, Amin MR, Azad AK. Production and Partial Characterization of Cellulase from Pseudomonas Isolates Obtained from Cow Dung and Municipal Solid Wastes. Bangladesh Journal of Microbiology 2013; 30: 11-16.
- [34]Khatiwada P, Ahmed J, Sohag MH, Islam K., Azad AK. Isolation, screening and characterization of cellulase producing bacterial isolates from municipal solid wastes and rice straw wastes. J Bioprocess Biotech 2016; 6: 280- 2.
- [35]Otajevwo FD, Aluyi HSA. Cultural conditions necessary for optimal cellulase yield by cellulolytic bacterial organisms as they relate to residual sugars released in broth medium. Modern Applied Science 2011; 5: 141.
- [36]Kushwaham A, Vipul V, Alpika V. Isolation & production of cellulase enzyme from bacteria isolated from agricultural fields in district Hardoi, Uttar Pradesh, India. Advances in Applied Science Research 2012; 3: 171-174.
- [37]Wen Z, Liao W, Chen S. Production of cellulase by Trichoderma reesei from dairy manure. Bioresource Technology 2005; 96 :4 91-499.